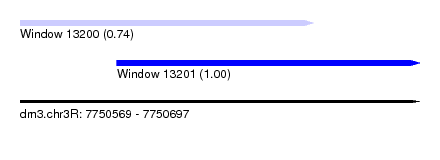
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,750,569 – 7,750,697 |
| Length | 128 |
| Max. P | 0.999836 |

| Location | 7,750,569 – 7,750,663 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 63.52 |
| Shannon entropy | 0.68604 |
| G+C content | 0.39643 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -10.70 |
| Energy contribution | -10.39 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.89 |
| Mean z-score | -0.62 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
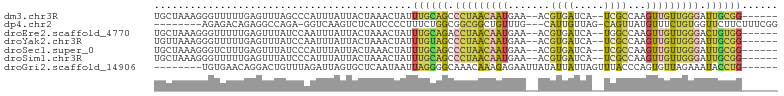
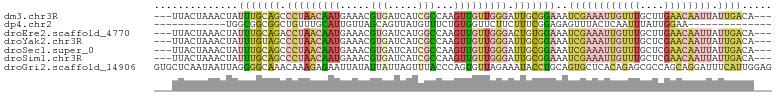
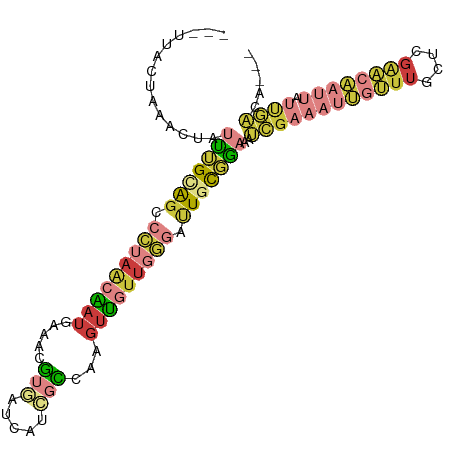
>dm3.chr3R 7750569 94 + 27905053 UGCUAAAGGGUUUUUGAGUUUAGCCCAUUUAUUACUAAACUAUUUGCAGCCCUAACAAUGAA--ACGUGAUCA--UCGCCAAGUUGUUGGGAUUGCGG------ .......(((((.........)))))..................((((..(((((((((...--..(((....--.)))...)))))))))..)))).------ ( -24.40, z-score = -1.46, R) >dp4.chr2 5434227 91 - 30794189 --------AGAGACAGAGGCCAGA-GGUCAAGUCUCAUCCCCUUUCUGGCGGCGGCUGUUUG---CAUUGUUAG-CAGUUAUGUUUCUGUGGUUCUUCUUUCGG --------((((((((..((((((-((...............))))))))..)(((((((.(---(...)).))-))))).)))))))................ ( -22.66, z-score = 0.71, R) >droEre2.scaffold_4770 3876649 94 - 17746568 UGCUAAAGGGUUUUUGAGUUUAUCCAAUUUAUUACUAAACUAUUUGCAGACCUAACAAUGAA--ACGUGAUCA--UGGCCAAGUUGUUGGGACUGUGG------ .......((((..........))))..................(..(((.(((((((((...--.(((....)--)).....))))))))).)))..)------ ( -20.30, z-score = -0.57, R) >droYak2.chr3R 12162805 94 - 28832112 UGUUAAAGGGUUUUUGAGUUUAUCCAAUUUAUUACUAAACUAUUUGUAGCCCUAACAAUGAA--ACGUGAUCA--UCGCCAAGUUGUUGGGAUUGCGG------ ((((..((((((....((((((.............))))))......)))))))))).....--.((..(((.--...((((....)))))))..)).------ ( -20.22, z-score = -0.89, R) >droSec1.super_0 6901602 94 - 21120651 UGCUAAAGGGUCUUUGAGUUUAUCCCAUUUAUUACUAAACUAUUUGCAGCCCUAACAAUGAA--ACGUGAUCA--UCGCCAAGUUGUUGGGAUUGCGG------ .......(((.............)))..................((((..(((((((((...--..(((....--.)))...)))))))))..)))).------ ( -21.62, z-score = -0.86, R) >droSim1.chr3R 13873551 94 - 27517382 UGCUAAAGGGUUUUUGAGUUUAUCCCAUUUAUUACUAAACUAUUUGCAGCCCUAACAAUGAA--ACGUGAUCA--UCGCCAAGUUGUUGGGAUUGCGG------ .......(((.............)))..................((((..(((((((((...--..(((....--.)))...)))))))))..)))).------ ( -21.62, z-score = -0.90, R) >droGri2.scaffold_14906 465263 90 + 14172833 --------UGUGAACAGGACUGUUUAGAUUAGUGCUCAAUAAUUAGGGGCAAACAAAGAGAAUUAUAUUAUUAGUUUACCCAGUGUUAGAAAUACCUG------ --------......((((((((..(((((((((((((.........))))...(.....).........)))))))))..))))..((....))))))------ ( -15.90, z-score = -0.35, R) >consensus UGCUAAAGGGUUUUUGAGUUUAUCCAAUUUAUUACUAAACUAUUUGCAGCCCUAACAAUGAA__ACGUGAUCA__UCGCCAAGUUGUUGGGAUUGCGG______ ............................................(((((.(((((((((.......((((.....))))...))))))))).)))))....... (-10.70 = -10.39 + -0.31)
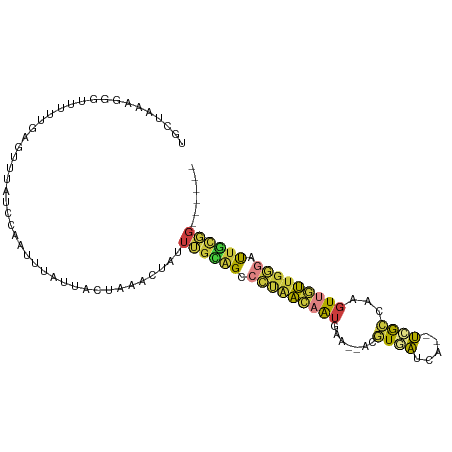
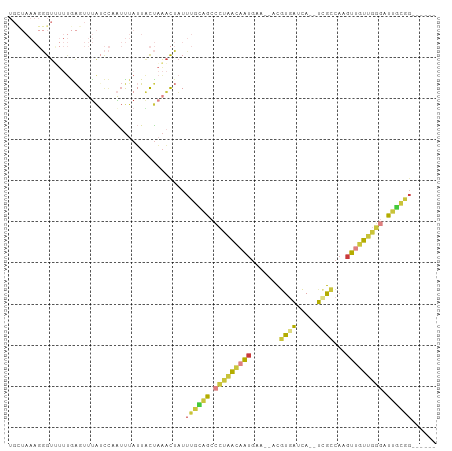
| Location | 7,750,600 – 7,750,697 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 61.12 |
| Shannon entropy | 0.77178 |
| G+C content | 0.38419 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -12.46 |
| Energy contribution | -14.74 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.53 |
| SVM RNA-class probability | 0.999836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7750600 97 + 27905053 ---UUACUAAACUAUUUGCAGCCCUAACAAUGAAACGUGAUCAUCGCCAAGUUGUUGGGAUUGCGGAAAUCGAAAUUGUUUGCUUGAACAAUUAUUGACA--- ---...........((((((..(((((((((.....(((.....)))...)))))))))..))))))..((((((((((((....)))))))).))))..--- ( -27.30, z-score = -3.40, R) >dp4.chr2 5434264 77 - 30794189 ------------UGGCGGCGGCUGUUUGCAUUGUUAGCAGUUAUGUUUCUGUGGUUCUUCUUUCGGAGAGUUUACUCAAUUUAUUGGAA-------------- ------------..((((..((...((((.......))))....))..))))..((((((....))))))...................-------------- ( -12.70, z-score = 1.15, R) >droEre2.scaffold_4770 3876680 97 - 17746568 ---UUACUAAACUAUUUGCAGACCUAACAAUGAAACGUGAUCAUGGCCAAGUUGUUGGGACUGUGGAAAUCGAAAUUGUUUGCUUGAACAAUUAUUGACA--- ---...........((..(((.(((((((((....(((....))).....))))))))).)))..))..((((((((((((....)))))))).))))..--- ( -27.10, z-score = -3.64, R) >droYak2.chr3R 12162836 97 - 28832112 ---UUACUAAACUAUUUGUAGCCCUAACAAUGAAACGUGAUCAUCGCCAAGUUGUUGGGAUUGCGGAAAUCGAAAUUGUUUGCUCGAACAAUUAUUGACA--- ---...........((((((..(((((((((.....(((.....)))...)))))))))..))))))..((((((((((((....)))))))).))))..--- ( -25.80, z-score = -2.87, R) >droSec1.super_0 6901633 97 - 21120651 ---UUACUAAACUAUUUGCAGCCCUAACAAUGAAACGUGAUCAUCGCCAAGUUGUUGGGAUUGCGGAAAUCGAAAUUGUUUGCUCGAACAAUUAUUGACA--- ---...........((((((..(((((((((.....(((.....)))...)))))))))..))))))..((((((((((((....)))))))).))))..--- ( -27.80, z-score = -3.42, R) >droSim1.chr3R 13873582 97 - 27517382 ---UUACUAAACUAUUUGCAGCCCUAACAAUGAAACGUGAUCAUCGCCAAGUUGUUGGGAUUGCGGAAAUCGAAAUUGUUUGCUCGAACAAUUAUUGACA--- ---...........((((((..(((((((((.....(((.....)))...)))))))))..))))))..((((((((((((....)))))))).))))..--- ( -27.80, z-score = -3.42, R) >droGri2.scaffold_14906 465286 103 + 14172833 GUGCUCAAUAAUUAGGGGCAAACAAAGAGAAUUAUAUUAUUAGUUUACCCAGUGUUAGAAAUACCUGCAGUGCUCACAGAGCGCCAGCAGGAUUUCAUUGGAG .(((((.........)))))........((((((......))))))..(((((....(((((.(((((.((((((...))))))..))))))))))))))).. ( -29.80, z-score = -3.11, R) >consensus ___UUACUAAACUAUUUGCAGCCCUAACAAUGAAACGUGAUCAUCGCCAAGUUGUUGGGAUUGCGGAAAUCGAAAUUGUUUGCUCGAACAAUUAUUGACA___ ..............(((((((.(((((((((.....(((.....)))...))))))))).)))))))..((((((((((((....)))))))).))))..... (-12.46 = -14.74 + 2.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:56 2011