
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,742,041 – 7,742,133 |
| Length | 92 |
| Max. P | 0.535996 |

| Location | 7,742,041 – 7,742,133 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.04 |
| Shannon entropy | 0.44859 |
| G+C content | 0.49655 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -15.36 |
| Energy contribution | -16.68 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
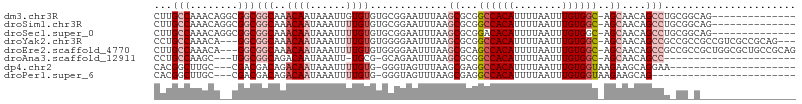
>dm3.chr3R 7742041 92 - 27905053 CUUGCCAAACAGGCGGCGGCAAACAAUAAAUUGUGUGUGCGGAAUUUAAGCGCGGCCACAUUUUAAUUUGUGGC-AGCAACAGCCUGCGGCAG-------------- ..((((...(((((..(.(((.((((....))))...))).).........((.((((((........))))))-.))....))))).)))).-------------- ( -34.60, z-score = -1.81, R) >droSim1.chr3R 13864948 92 + 27517382 CUUGCCAAACAGGCGGCGGCAAACAAUAAAUUUUGUGUGCGGAAUUUAAGCGCGGCCACAUUUUAAUUUGUGGC-AGCAACAGCCUGCGGCAG-------------- ..((((...(((((..(.(((.((((......)))).))).).........((.((((((........))))))-.))....))))).)))).-------------- ( -35.80, z-score = -2.62, R) >droSec1.super_0 6892967 92 + 21120651 CUUGCCAAACAGGCGGCGGCAAACAAUAAAUUUUGUGUGCGGAAUUUAAGCGCGGACACAUUUUAAUUUGUGGC-AGCAACAGCCUGCGGCAG-------------- ..((((...(((((.((.((.....(((((((.(((((.(((........).)).)))))....))))))).))-.))....))))).)))).-------------- ( -30.40, z-score = -1.60, R) >droYak2.chr3R 12153996 100 + 28832112 CCUGCCAAACA---GGCGGCAAACAAUAAAUUUUGUGUGGGGAAUUUAAGCGCGGCCACAUUUUAAUUUGUGGC-AGCAACAGCCGCCGCCGCCGUCGCCGCAG--- .((((......---((((((..((((......))))..((((.......((...((((((........))))))-.)).....)).))))))))......))))--- ( -37.60, z-score = -1.45, R) >droEre2.scaffold_4770 3867878 103 + 17746568 CUUGCCAAACA---GGCGGCAAACAAUAAAUUUUGUGUGGGGAAUUUAAGCGCAGCCACAUUUUAAUUUGUGGC-AGCAACAGCCGCCGCCGCUGGCGCUGCCGCAG ...((((....---((((((......((((((((......))))))))...((.((((((........))))))-.)).......))))))..))))((....)).. ( -37.60, z-score = -0.74, R) >droAna3.scaffold_12911 2409411 79 - 5364042 CCUGCCAAGC---UGGCGGCAGACAAUAAAUU-UGCG-GCAGAAUUUAAGCGCGGCCACAUUUUAAUUUGUGGC-AGCAACAGCC---------------------- ........((---((((.(((((.......))-))).-))...........((.((((((........))))))-.))..)))).---------------------- ( -27.50, z-score = -1.85, R) >dp4.chr2 5424053 82 + 30794189 CACGGCUUGC---CGACGACAGACAAUAAAUUUUGUG-GGGUAGUUUAAGCGAGGCCACAUUUUAAUUUGUGGUAAGAAGCAGGAA--------------------- ..(((....)---)).(.(((((........))))).-)..........((...((((((........)))))).....)).....--------------------- ( -18.90, z-score = -0.91, R) >droPer1.super_6 5441666 79 + 6141320 CACGGCUUGC---CGACGACAGACAAUAAAUUUUGUG-GGGUAGUUUAAGCGAGGCCACAUUUUAAUUUGUGGUAAGAAGCAG------------------------ ..(((....)---)).(.(((((........))))).-)..........((...((((((........)))))).....))..------------------------ ( -18.90, z-score = -1.22, R) >consensus CUUGCCAAACA__CGGCGGCAAACAAUAAAUUUUGUGUGCGGAAUUUAAGCGCGGCCACAUUUUAAUUUGUGGC_AGCAACAGCCU_CG_C________________ ...(((........)))(((..((((......)))).............((...((((((........))))))..))....)))...................... (-15.36 = -16.68 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:52 2011