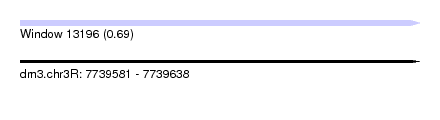
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,739,581 – 7,739,638 |
| Length | 57 |
| Max. P | 0.685612 |

| Location | 7,739,581 – 7,739,638 |
|---|---|
| Length | 57 |
| Sequences | 10 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 70.53 |
| Shannon entropy | 0.55631 |
| G+C content | 0.32344 |
| Mean single sequence MFE | -5.64 |
| Consensus MFE | -3.68 |
| Energy contribution | -3.47 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.62 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.685612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7739581 57 + 27905053 GUAAAAACUUUAAGUCUAAUGACUUUUAUAGAUACUCCA-CAAUAAGUCCGUGCCCAU------------- (((...((((.(((((....)))))..............-....))))...)))....------------- ( -5.01, z-score = -0.74, R) >droSim1.chr3R 13862526 57 - 27517382 GUAAAAACUUUAAGUCUAAUGACUUUUAUAGAUACUCCA-CAAUAAGUCCGUGCCCAU------------- (((...((((.(((((....)))))..............-....))))...)))....------------- ( -5.01, z-score = -0.74, R) >droSec1.super_0 6890532 57 - 21120651 GUAAAAACUUUAAGUCUAAUGACUUUUAUAGAUACUCCA-CAAUAAGUCCGUGCCCAU------------- (((...((((.(((((....)))))..............-....))))...)))....------------- ( -5.01, z-score = -0.74, R) >droYak2.chr3R 12151551 57 - 28832112 GUAAAAACUUUAAGUCUAAUGACUUUUAUAGACACUUCA-CAAUAAGUCAAUGCCCAU------------- (((........(((((....))))).....(((......-......)))..)))....------------- ( -5.40, z-score = -0.82, R) >droEre2.scaffold_4770 3865232 57 - 17746568 GUAAAAACUUUAAGUCUAAUGACUUUUAUAGACACUCCA-CAAUAAGUCUAUGCUCAU------------- ...........(((((....))))).(((((((......-......))))))).....------------- ( -8.90, z-score = -2.27, R) >droAna3.scaffold_12911 2407178 52 + 5364042 GCAAAAACUUUAAGUCUAAUGACUUUUGUAGAUACUUCG-CAGUAAGCCCGAC------------------ ((....((...(((((....)))))..)).((....)))-)............------------------ ( -7.40, z-score = -0.57, R) >dp4.chr2 5420927 71 - 30794189 GUUAAAACUUUAAGUCUAAUGACUUUUAUAGAUACUUCAACCAUAAGUCCAUCUCUCCAUUCAUUGCCCCU .......((..(((((....)))))....))........................................ ( -4.70, z-score = -0.67, R) >droPer1.super_6 5438545 71 - 6141320 GUUAAAACUUUAAGUCUAAUGACUUUUAUAGAUACUUCAACCAUAAGUCCAUCUCUCCAUUCGUUGCCCCU .....(((...(((((....)))))....((((((((.......))))..))))........)))...... ( -5.20, z-score = -0.72, R) >droVir3.scaffold_13047 3800598 57 + 19223366 UAGUAAACUUUAAGUUUAAUAUCUUUUAUGGAUACUUAA---GAAAAUGUGCGUGUGUGU----------- ..(((...(((((((.....((((.....))))))))))---)......)))........----------- ( -5.00, z-score = 0.70, R) >droMoj3.scaffold_6540 20802569 50 + 34148556 CAGUAAACUUAAAGUUUAAUGUCUUUUAUGGAUGCUUGA---GAAAUUCUGUG------------------ (((....(((((.((.....((((.....))))))))))---).....)))..------------------ ( -4.80, z-score = 0.34, R) >consensus GUAAAAACUUUAAGUCUAAUGACUUUUAUAGAUACUUCA_CAAUAAGUCCGUGCCCAU_____________ .......((..(((((....)))))....))........................................ ( -3.68 = -3.47 + -0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:52 2011