| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,692,898 – 7,693,007 |
| Length | 109 |
| Max. P | 0.633753 |

| Location | 7,692,898 – 7,693,007 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Shannon entropy | 0.47339 |
| G+C content | 0.41725 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -11.53 |
| Energy contribution | -12.01 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7692898 109 + 27905053 AAACUUUUAAUUUGUGCGU-AGCUAGCGAAAGUAUAUGAAAAAUGGCACGAAAACAAAAGUUC---CCCAUAAAAAGUAGACGCA--UGGAUGCAGGUCCAUGGAC-ACCAUGGAC--- ...((....(((..(((((-.(((.((....)).........((((...(((........)))---.))))....)))..)))))--..)))..))((((((((..-.))))))))--- ( -28.70, z-score = -2.66, R) >droSim1.chr3R 13813764 109 - 27517382 AAACUUUUAAUUUGUGCGU-AGCUAGCGAAAGUAUAUGAAAAAUGGCACGAAAACAAAAGUUG---CCCAUAAAAAGUAGACGCA--UGGAUGCAGGUCCAUGGAC-ACCAUGGAC--- ...((....(((..(((((-.(((.((....))...........(((((..........).))---)).......)))..)))))--..)))..))((((((((..-.))))))))--- ( -30.10, z-score = -2.97, R) >droSec1.super_0 6843901 109 - 21120651 AAACUUUUAAUUUGUGCGU-AGCUAGCGAAAGUAUAUGAAAAAUGGCACGAAAACAAAAGUUG---CCCAUAAAAAGUAGACGCA--UGGAUGCAGGUCCAUGGAC-ACCAUGGAC--- ...((....(((..(((((-.(((.((....))...........(((((..........).))---)).......)))..)))))--..)))..))((((((((..-.))))))))--- ( -30.10, z-score = -2.97, R) >droYak2.chr3R 12103293 112 - 28832112 AAACUUUUAAUUUGUGCGU-AGCUAGCGAAAGUAUAUGAAAAAUGGCACGAAAACAAAAGUUC---CGUAUAAAAAGUAGACGCA--CGGAUGCUGGUCCAUGGAC-ACCAGGAACACC ..........(((((.(((-.((((((....))...(....).)))))))...))))).((((---(........((((..(...--.)..)))).(((....)))-....)))))... ( -26.60, z-score = -1.61, R) >droAna3.scaffold_12911 2346973 108 + 5364042 AAACUUUUAAUUUGUGCGU-AGCUAGCGAAAGUAUAUGAAAAAUGGCACGAAAACAAAAGUUG---CCAGCAAA--GUUGAUGCA--UGGAUGCUGGUCUUGGGAGGACCGGGCAC--- .........(((..(((((-((((.((....))..........((((((..........).))---)))....)--))).)))))--..))).(((((((.....)))))))....--- ( -30.50, z-score = -1.89, R) >dp4.chr2 5366766 113 - 30794189 AAACUUUUAAAUUGUGCGUCACCCAACGAAAGUAUAUGAAAAAUGGCACGAAAACAAAAGUUUUUUCCAUCAAAGGAGCGGCAGGACCUGCUGCUGGCAACAGGGAAUGCAGG------ ((((((((...((((((.(((....((....))...)))......))))))....))))))))(((((........((((((((...))))))))(....).)))))......------ ( -29.70, z-score = -1.33, R) >droPer1.super_6 5384594 113 - 6141320 AAACUUUUAAAUUGUGCGUCACCCAACGAAAGUAUAUGAAAAAUGGCACGAAAACAAAAGUUUUUUCCAUCAAAGGAGCGGCAGGACCUGCUGCUGGUAACAGGGAAUGCAGG------ ..............(((((..(((.((....))...((....((((...((((((....)))))).))))......((((((((...)))))))).....))))).)))))..------ ( -28.40, z-score = -1.10, R) >droWil1.scaffold_181130 900035 97 - 16660200 AAACUUUUAAUUUGUGCGU-AGC----GAAAGUAAAUGAAAAAUGGCACAUAAACAAAAGUUA---CCAGCAAAUAGCC--CGCG--UUGCCAGAGCCCAGAAGAC-GCC--------- .(((((((....(((((..-.((----....))...(....)...))))).....))))))).---...((.....)).--.(((--((..(........)..)))-)).--------- ( -18.00, z-score = -0.77, R) >consensus AAACUUUUAAUUUGUGCGU_AGCUAGCGAAAGUAUAUGAAAAAUGGCACGAAAACAAAAGUUG___CCAACAAAAAGUAGACGCA__UGGAUGCAGGUCCAUGGAC_ACCAGG_AC___ .(((((((..(((((((........((....)).(((.....))))))))))...)))))))......................................................... (-11.53 = -12.01 + 0.48)

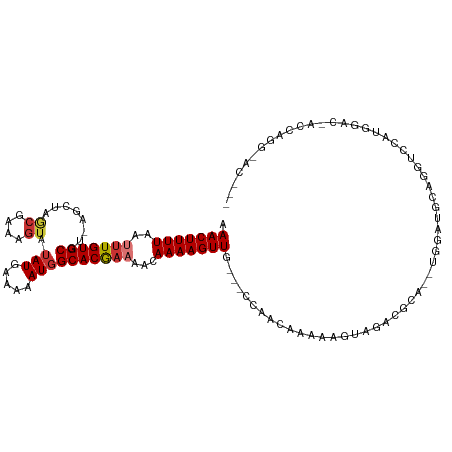

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:44 2011