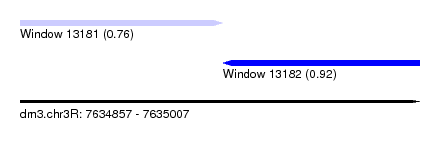
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,634,857 – 7,635,007 |
| Length | 150 |
| Max. P | 0.920745 |

| Location | 7,634,857 – 7,634,933 |
|---|---|
| Length | 76 |
| Sequences | 7 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 68.90 |
| Shannon entropy | 0.55108 |
| G+C content | 0.36368 |
| Mean single sequence MFE | -15.18 |
| Consensus MFE | -4.48 |
| Energy contribution | -5.62 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
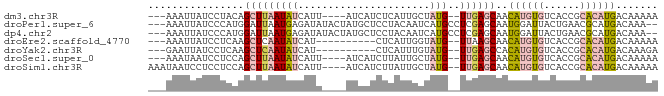
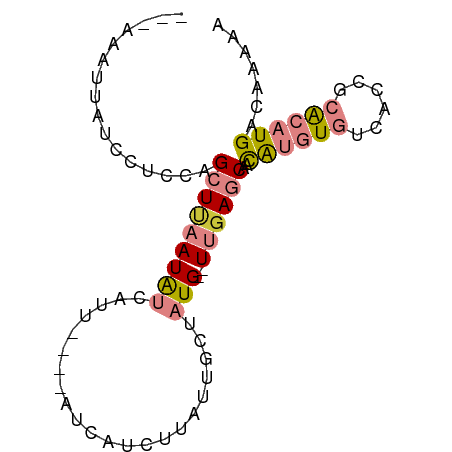
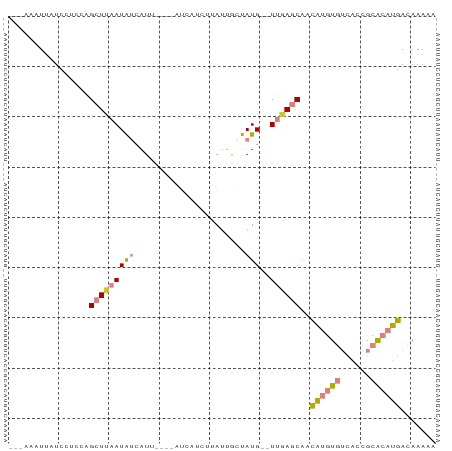
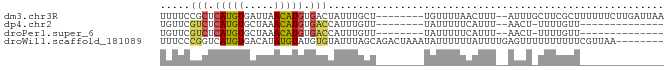
>dm3.chr3R 7634857 76 + 27905053 ---AAAUUAUCCUACAGCUUAAUAUCAUU----AUCAUCUCAUUGCUAUG--UUGAGCAACAUGUGUCACCGCACAUGACAAAAA ---.............(((((((((((..----..........))..)))--))))))..(((((((....)))))))....... ( -15.50, z-score = -2.62, R) >droPer1.super_6 1201945 80 + 6141320 ---AAAUUAUCCCAUGGAUUAAUGAGAUAUACUAUGCUCCUACAAUCAUGCCUCGAGCAAUGGAUUACUGAACGCAUGACAAA-- ---....((((.(((......))).))))...(((((......((((.(((.....)))...)))).......))))).....-- ( -11.02, z-score = -0.05, R) >dp4.chr2 5069453 80 - 30794189 ---AAAUUAUCCCAUGGAUUAAUGAGAUAUACUAUGCUCCUACAAUCAUGCCUCGAGCAAUGGAUUACUGAACGCAUGACAAA-- ---....((((.(((......))).))))...(((((......((((.(((.....)))...)))).......))))).....-- ( -11.02, z-score = -0.05, R) >droEre2.scaffold_4770 3764737 70 - 17746568 ---AAAUUAUCCUCAAGCUCAAUAUCAU----------CUCAUUGGUAUG--UUAAGCAACAUGUGUCACCGCACAUGACAAAAA ---.............(((..((((((.----------.....)))))).--...)))..(((((((....)))))))....... ( -15.40, z-score = -3.46, R) >droYak2.chr3R 12048890 70 - 28832112 ---GAAUUAUCCUCAAGCUCAAUAUCAU----------CUCAUUUGUAUG--UUGAGCCACAUGUGUCACCGCACAUGACAAAGA ---.............(((((((((((.----------......)).)))--))))))..(((((((....)))))))....... ( -19.50, z-score = -3.43, R) >droSec1.super_0 6790865 76 - 21120651 ---AAAUAAUCCUCCAGCUUAAUAUCAUU----AUCAUCUUAUUGCUAUG--UUGAGCAACAUGUGUCACCGCACAUGACAAAAA ---.............(((((((((((.(----(......)).))..)))--))))))..(((((((....)))))))....... ( -16.90, z-score = -3.52, R) >droSim1.chr3R 13760503 79 - 27517382 AAAUAAUCCUCCUCCAGCUUAAUAUCAUU----AUCAUCUUAUUGCUAUG--UUGAGCAACAUGUGUCACCGCACAUGACAAAAA ................(((((((((((.(----(......)).))..)))--))))))..(((((((....)))))))....... ( -16.90, z-score = -3.78, R) >consensus ___AAAUUAUCCUCCAGCUUAAUAUCAUU____AUCAUCUUAUUGCUAUG__UUGAGCAACAUGUGUCACCGCACAUGACAAAAA ................((((((...(((...................)))..))))))..((((((......))))))....... ( -4.48 = -5.62 + 1.15)
| Location | 7,634,933 – 7,635,007 |
|---|---|
| Length | 74 |
| Sequences | 4 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 58.69 |
| Shannon entropy | 0.59544 |
| G+C content | 0.28418 |
| Mean single sequence MFE | -8.43 |
| Consensus MFE | -4.72 |
| Energy contribution | -4.85 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
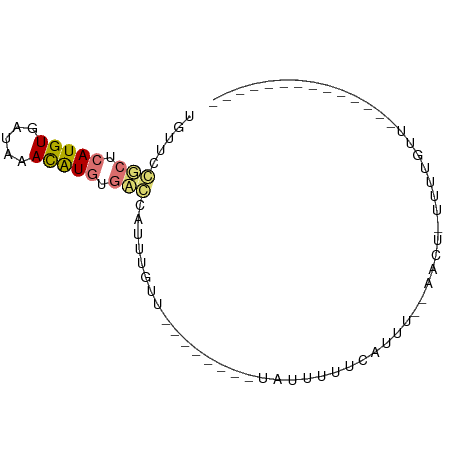
>dm3.chr3R 7634933 74 - 27905053 UUUUCCGCUCAUGUGAUUAACAUGUGACUAUUUGCU--------UGUUUUAACUUU--AUUUGCUUCGCUUUUUUCUUGAUUAA ......(((((((((.....)))))))......((.--------.((.........--))..))...))............... ( -6.60, z-score = -0.28, R) >dp4.chr2 5069533 59 + 30794189 UGUUCGUCUCAUGUGCUAAACAUGUGACCAUUUGUU--------UAUUUUUCAUUU--AACU-UUUUGUU-------------- .....(((.(((((.....))))).)))........--------............--....-.......-------------- ( -9.30, z-score = -2.14, R) >droPer1.super_6 1202025 59 - 6141320 UGUUCGUCUCAUGUGCUAAACAUGUGACCAUUUGUU--------UAUUUUUCAUUU--AACU-UUUUGUU-------------- .....(((.(((((.....))))).)))........--------............--....-.......-------------- ( -9.30, z-score = -2.14, R) >droWil1.scaffold_181089 6641979 76 - 12369635 UUUCCCGGUCAUGUGACAUAUGUAUGUGUAUUUAGCAGACUAAAUAUUUUUUAUUUUGAGUUUUUUUUUUCGUUAA-------- ......((((.(((.(((((....))))).....)))))))................(((........))).....-------- ( -8.50, z-score = -0.17, R) >consensus UGUUCCGCUCAUGUGAUAAACAUGUGACCAUUUGUU________UAUUUUUCAUUU__AACU_UUUUGUU______________ .....(((.(((((.....))))).)))........................................................ ( -4.72 = -4.85 + 0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:40 2011