| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,125,468 – 7,125,560 |
| Length | 92 |
| Max. P | 0.728606 |

| Location | 7,125,468 – 7,125,560 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 61.03 |
| Shannon entropy | 0.73967 |
| G+C content | 0.44764 |
| Mean single sequence MFE | -14.72 |
| Consensus MFE | -4.89 |
| Energy contribution | -4.28 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
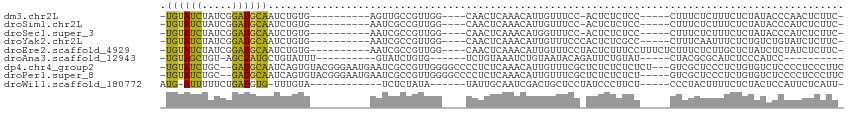
>dm3.chr2L 7125468 92 + 23011544 -UGUAUCUAUCGGAUGCAAUCUGUG----------AGUUGCCGUUGG----CAACUCAAACAUUGUUUCC-ACUCUCUCC-----CUUUCUCUUUCUCUAUACCCAACUCUUC- -..........(((.(((((...((----------(((((((...))----)))))))...))))).)))-.........-----............................- ( -23.10, z-score = -5.84, R) >droSim1.chr2L 6915028 92 + 22036055 -UGUAUCUAUCGGAUGCAAUCUGUG----------AAUCGCCGUUGG----CAACUCAAACAUUGUUUCC-ACUCUCUCC-----CUUUCUCUUUCUCUAUACCCAUCUCUUC- -..........(((.(((((...((----------(...(((...))----)...)))...))))).)))-.........-----............................- ( -13.10, z-score = -2.40, R) >droSec1.super_3 2649216 92 + 7220098 -UGUAUCUAUCGGAUGCAAUCUGUG----------AAUCGCCGUUGG----CAACUCAAACAUGGUUUCC-ACUCUCUCC-----CUUUCUCUUUCUCUAUACCCAUCUCUUC- -((((((.....))))))....(((----------....(((((.(.----...)......)))))...)-)).......-----............................- ( -10.90, z-score = -0.51, R) >droYak2.chr2L 16543554 93 - 22324452 -UGUAUCUAUCGGAUGCAAUCUGUG----------AAUCGCCGUUGG----CAACUCAAACAUUGUUUCCCACUCUCGCC-----CUUUCAAUUUCUCUGUCUGUAUCUCUUC- -..........(((.(((((...((----------(...(((...))----)...)))...))))).)))..........-----............................- ( -12.20, z-score = -0.59, R) >droEre2.scaffold_4929 16040948 98 + 26641161 -UGUAUCUAUCGGAUGCAAUCUGUG----------AAUCGCCGUUGG----CAACUCAAACAUUGUUUCCUACUCUUUCCUUUCUCUUUCUCUUGCUCUAUCUCUAUCUCUUC- -..........(((.(((((...((----------(...(((...))----)...)))...))))).)))...........................................- ( -12.60, z-score = -2.26, R) >droAna3.scaffold_12943 975489 81 - 5039921 -UGUAGCUGU-AGCUAUGCUGUAUUU----------GUAUCUGUG------UCUGUAAAUCUGUAAUACAGAUUCUGUAU-----CUACGCGCAUCUCCCAUCC---------- -.(((((...-.)))))((((((...----------((((..(.(------((((((.........))))))).).))))-----.)))).))...........---------- ( -15.20, z-score = -0.92, R) >dp4.chr4_group2 751013 108 + 1235136 -UGUAUCUGC--GAUGCAAUCAGUGUACGGGAAUGAAUCGCCGUUGGGGCCCCUCUCAAACAUUGUUUCGCUCUCUCUCUCU---GUCGCUCCCUCUGUGUCUCCCCUCCCUUC -((((((...--))))))..........((((..((..(((.(..(((((..(...........(.....)...........---)..)))))..).)))..))...))))... ( -20.55, z-score = 0.07, R) >droPer1.super_8 820178 106 - 3966273 -UGUAUCUGC--GAUGCAAUCAGUGUACGGGAAUGAAUCGCCGUUGGGGCCCCUCUCAAACAUUGUUUCGCUCUCUCUCU-----GUCGCUCCCUCUGUGUCUCCCCUCCCUUC -((((((...--))))))..........((((..((..(((.(..(((((..(...........................-----)..)))))..).)))..))...))))... ( -20.33, z-score = 0.25, R) >droWil1.scaffold_180772 3189822 88 - 8906247 AUG-AUUUUUCUGAUGUG-UUUGUA------------UCUCUAUA------UAUUGCAAUCGACUGCUCCUAUCCCUUCU-----CCCUACUUUUCUCUACUCCAUUCUCAUU- (((-(.......((((((-(.....------------.....)))------))))(((......))).............-----.......................)))).- ( -4.50, z-score = -0.32, R) >consensus _UGUAUCUAUCGGAUGCAAUCUGUG__________AAUCGCCGUUGG____CAACUCAAACAUUGUUUCC_ACUCUCUCC_____CUUUCUCUUUCUCUAUCCCCAUCUCUUC_ .((((((.....))))))................................................................................................ ( -4.89 = -4.28 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:56 2011