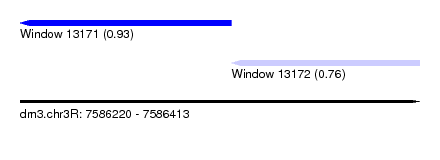
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,586,220 – 7,586,413 |
| Length | 193 |
| Max. P | 0.926010 |

| Location | 7,586,220 – 7,586,322 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 75.80 |
| Shannon entropy | 0.46470 |
| G+C content | 0.38024 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -9.14 |
| Energy contribution | -10.25 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.926010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
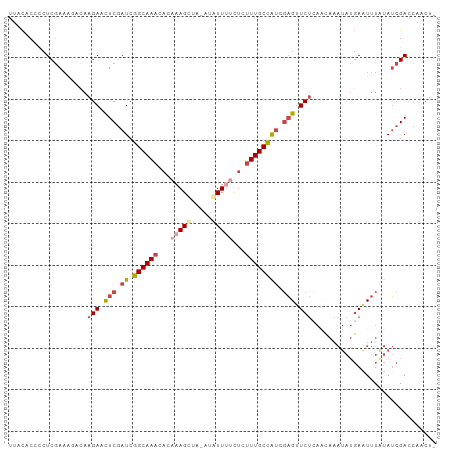
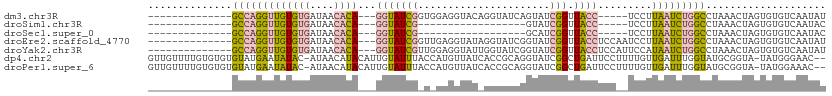
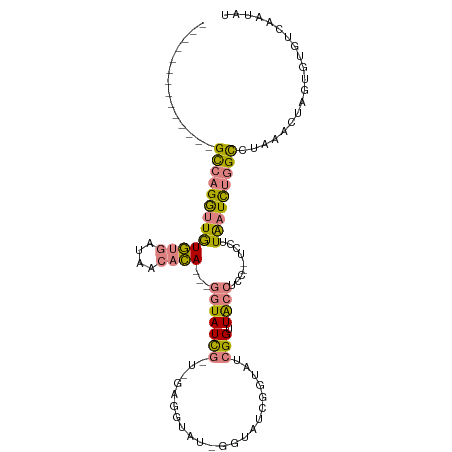
>dm3.chr3R 7586220 102 - 27905053 UUACACCCCUCGAAAGACAAGAACUCGAUCGGCAAACACAUAGCUA-AUGUUUUCUCUUUGCCAUCGAGAUCUCAGCAAAUAUGAGUUUGUAUCGACCAACUA .........((((...(((((..((((((.((((((.....(((..-..))).....))))))))))))..((((.......))))))))).))))....... ( -25.70, z-score = -3.52, R) >droSim1.chr3R 13708334 101 + 27517382 UUACACCCCUCGAAAGACAAGAACUCGAUCGGCAAACACAAAGCUA-AUAUUUUCUCUUUGCCAUGGAGUUCUCAAUAAAUAUGAAUUUAUAUCGACCGACU- .........((((......(((((((.((.((((((.(.((((...-...)))).).)))))))).)))))))..((((((....)))))).))))......- ( -20.70, z-score = -3.00, R) >droSec1.super_0 6741070 101 + 21120651 UUACACCCCUCGAAAGACAAGAACUCGAUCGGCAAACACAAAGCUA-AUAUUUUCUCUUUGCCAUGGAGUUCUCAAUAAAUAUGAAUUUAUAUCGACCGACU- .........((((......(((((((.((.((((((.(.((((...-...)))).).)))))))).)))))))..((((((....)))))).))))......- ( -20.70, z-score = -3.00, R) >droEre2.scaffold_4770 3715985 96 + 17746568 UUACACCCCUCGA-----AAGAUUUCGAUCGGCAAACACCAAGCUA-AUGUUUUCUUUUUGCCAUCGAGUUCUGAACAAAUAUGAAUUUAUAUCGACCAGCU- .........((((-----.(((.((((((.((((((....((((..-..))))....)))))))))))).)))......(((......))).))))......- ( -19.70, z-score = -2.60, R) >droYak2.chr3R 11999844 101 + 28832112 UUGCACCUCUCAAAGACCAAGAUCUCCAUCGGCAAACACAAAGCCA-AUGUUUUCUUUUUGCCAUCGAGUUCUCAACAAAUAUGAAUUUAUAUCGACUAACU- ...................(((.(((.((.((((((.(.(((((..-..))))).).)))))))).))).))).............................- ( -13.50, z-score = -0.83, R) >droAna3.scaffold_13340 14067065 90 - 23697760 AUAUCCACAUCGAAAUCCAAGAACUCGAUCUGCAAUAAUAAAGUUGUUUAUUGAUAUUUUGCGGCUUUGUUC------------AGUUUCCAGCGAUCGAAU- ........................((((((.((((((((((((((((..(.......)..))))))))))).------------.)))....))))))))..- ( -15.70, z-score = -0.44, R) >consensus UUACACCCCUCGAAAGACAAGAACUCGAUCGGCAAACACAAAGCUA_AUAUUUUCUCUUUGCCAUCGAGUUCUCAACAAAUAUGAAUUUAUAUCGACCAACU_ ...................(((.(((.((.((((((...(((((.....)))))...)))))))).))).))).............................. ( -9.14 = -10.25 + 1.11)


| Location | 7,586,322 – 7,586,413 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 59.93 |
| Shannon entropy | 0.69829 |
| G+C content | 0.42402 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -7.53 |
| Energy contribution | -7.96 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.759145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7586322 91 - 27905053 --------------GCCAGGUUGUGUGAUAACACA---GGUAUCGGUGGAGGUACAGGUAUCAGUAUCGGUUACC-----UCCUUAAUCUGGCCUAAACUAGUGUGUCAAUAU --------------((((((((((((....)))).---.........(((((((..((((....))))...))))-----)))..)))))))).................... ( -28.40, z-score = -1.78, R) >droSim1.chr3R 13708435 73 + 27517382 --------------GCCAGGUUGUGUGAUAACACA---GGUAUCG------------------GUAUCGGUUACC-----UCCUUAAUCUGGCCUAAACUAGUGUGUCAAUAC --------------((((((((((((....))))(---(((((((------------------....))).))))-----)....))))))))........((((....)))) ( -21.80, z-score = -2.05, R) >droSec1.super_0 6741171 73 + 21120651 --------------GCCAGGUUGUGUGAUAACACA---GGUAUCG------------------GCAUCGGUUACC-----UCCUUAAUCUGGCCUAAACUAGUGUGUCAAUAC --------------((((((((((((....))))(---(((((((------------------....))).))))-----)....))))))))........((((....)))) ( -21.80, z-score = -1.97, R) >droEre2.scaffold_4770 3716081 96 + 17746568 --------------GCCAGGUUGUGUGAUAACACA---GGUAUCGGUUGAGGUAUAGGUAUCGGUAUCGGUUACCUCCAAUCCUUAAUCUGGCCUAAACUAGUGUGUCAAUAU --------------((((((((((((....)))).---......((((((((((..((((....))))...))))).)))))...)))))))).................... ( -27.80, z-score = -1.55, R) >droYak2.chr3R 11999945 96 + 28832112 --------------GCCAGGUUGUGUGAUAACACA---GGUAUCGUUGGAGGUAUUGGUAUCGGUAUCGGUUACCUCCAUUCCAUAAUCUGGCCUAAACUAGUGUGUCAAUAU --------------(((((((((((.((((.....---..))))..((((((((((((((....)))))).))))))))...))))))))))).................... ( -32.90, z-score = -3.05, R) >dp4.chr2 5022934 109 + 30794189 GUUGUUUUGUGUGUGUAUGAAUAUAC-AUAACAUACAUUGUAUUUACCAUGUUAUCACCGCAGGUAUCGGCUGAUUCCUUUUGUUGAUUUGGUAUGCGGUA-UAUGGGAAC-- ((((((.(((((((......))))))-).)))).))..........((((((....((((((...((((((.((.....)).))))))......)))))))-)))))....-- ( -29.40, z-score = -1.33, R) >droPer1.super_6 1152888 109 - 6141320 GUUGUUUUGUGUGUGUAUGAAUAUAC-AUAACAUACAUUGUAUUUACCAUGUUAUCACCGCAGGUAUCGGCUGAUUCCUUUUGUUGAUUUGGUAUGCGGUA-UAUGGAAAC-- ((((((.(((((((......))))))-).)))).))..........((((((....((((((...((((((.((.....)).))))))......)))))))-)))))....-- ( -29.20, z-score = -1.51, R) >consensus ______________GCCAGGUUGUGUGAUAACACA___GGUAUCG_U_GAGGUAU_GGUAUCGGUAUCGGUUACCUCC__UCCUUAAUCUGGCCUAAACUAGUGUGUCAAUAU ..............((((((((((((....))))....(((((....................))))).................)))))))).................... ( -7.53 = -7.96 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:31 2011