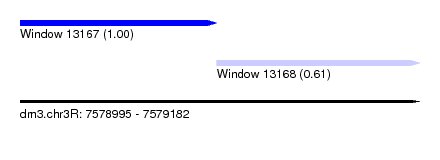
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,578,995 – 7,579,182 |
| Length | 187 |
| Max. P | 0.995904 |

| Location | 7,578,995 – 7,579,087 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.03 |
| Shannon entropy | 0.40922 |
| G+C content | 0.66861 |
| Mean single sequence MFE | -49.28 |
| Consensus MFE | -27.43 |
| Energy contribution | -28.84 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.995904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
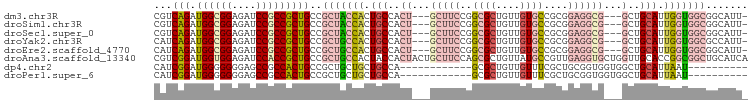
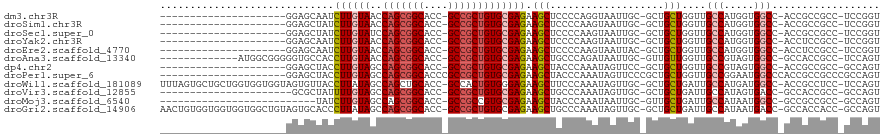
>dm3.chr3R 7578995 92 + 27905053 CGUCAGAUGGCGGAGAUCCGCCGCUGCCGCUACCACUGCCACU---GCUUCCGGCGCUGUUGUGCCGCGGAGGCG---GCUGCAUUGGUGGCGGCAUU- ....((.((((((....))))))))((((((((((.(((..((---(((((((((((....)))))..)))))))---)..))).))))))))))...- ( -58.60, z-score = -5.04, R) >droSim1.chr3R 13700926 92 - 27517382 CGUCAGAUGGCGGAGAUCCGCCGCUGCCGCUACCACUGCCACU---GCUUCCGGCGCUGUUGUGCCGCGGAGGCG---GCUGCAUUGGUGGCGGCAUU- ....((.((((((....))))))))((((((((((.(((..((---(((((((((((....)))))..)))))))---)..))).))))))))))...- ( -58.60, z-score = -5.04, R) >droSec1.super_0 6733717 92 - 21120651 CGUCAGAUGGCGGAGAUCCGCCGCUGCCGCUACCACUGCCACU---GCUUCCGGCGCUGUUGUGCCGCGGAGGCG---GCUGCAUUGGUGGCGGCAUU- ....((.((((((....))))))))((((((((((.(((..((---(((((((((((....)))))..)))))))---)..))).))))))))))...- ( -58.60, z-score = -5.04, R) >droYak2.chr3R 11992361 92 - 28832112 CAUCAGAUGGCGGAGAUCCGCCGCUGCCGCUGCCACUGCCACU---GCUUCCGGCGCUGUUGUGCCGCGGAGGCG---GCUGCAUUGGUGGCGCCAUU- ...(((.((((((....))))))))).(((..(((.(((..((---(((((((((((....)))))..)))))))---)..))).)))..))).....- ( -52.10, z-score = -3.07, R) >droEre2.scaffold_4770 3708484 92 - 17746568 CAUCAGAUGGCGGAGAUCCGCCGCUGCCGCUGCCACUGCCACU---GCUUCCGGCGCUGUUGUGCCGCGGAGGCG---GCUGCAUUGGUGGCGGCAUU- ....((.((((((....))))))))(((((..(((.(((..((---(((((((((((....)))))..)))))))---)..))).)))..)))))...- ( -57.70, z-score = -4.49, R) >droAna3.scaffold_13340 14060089 99 + 23697760 CGUCGGAUGGUGGAGAUCCACCGCUGCCGCUGCCACUACCACUACUGCUUCCAGCGCUGUUAUGCCGUUGAGGUGCUGGUUGCACCGGCGGCUGCAUCA .....((((((((....)))))((.(((((((.............(((..((((((((.(((......)))))))))))..))).))))))).))))). ( -41.24, z-score = -1.18, R) >dp4.chr2 5015948 77 - 30794189 CAUCGGAUGGGGGGGAGCCGCCACUGCCGCUGCUGCUGCCA------------GCGCUGUUGUUUCGCUGCGGUGGUGGCUGCAUUAAU---------- ...(((.(((.((....)).)))...))).(((.((..(((------------.(((.((......)).))).)))..)).))).....---------- ( -33.70, z-score = -0.32, R) >droPer1.super_6 1145882 77 + 6141320 CAUCGGAUGGGGGGGAGCCGCCACUGCCGCUGCUGCUGCCA------------GCGCUGUUGUUUCGCUGCGGUGGUGGCUGCAUUAAU---------- ...(((.(((.((....)).)))...))).(((.((..(((------------.(((.((......)).))).)))..)).))).....---------- ( -33.70, z-score = -0.32, R) >consensus CAUCAGAUGGCGGAGAUCCGCCGCUGCCGCUGCCACUGCCACU___GCUUCCGGCGCUGUUGUGCCGCGGAGGCG___GCUGCAUUGGUGGCGGCAUU_ ...(((.((((((....)))))))))..(((((((.(((.......(((((..((((....))))....))))).......))).)))))))....... (-27.43 = -28.84 + 1.41)
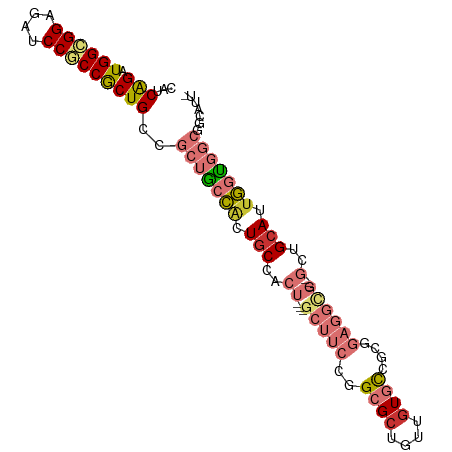
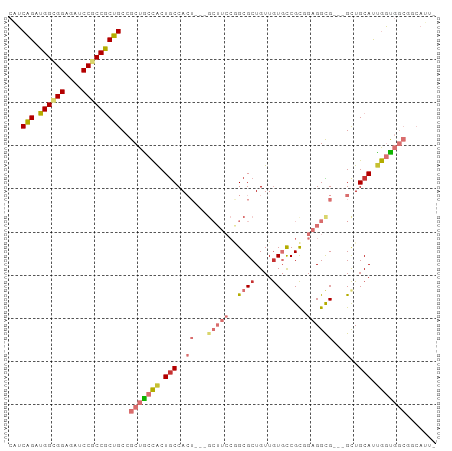
| Location | 7,579,087 – 7,579,182 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.81 |
| Shannon entropy | 0.38342 |
| G+C content | 0.62609 |
| Mean single sequence MFE | -42.07 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.41 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
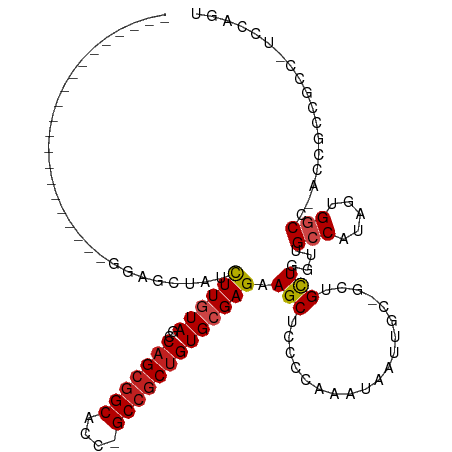
>dm3.chr3R 7579087 95 + 27905053 ---------------------GGAGCAAUCUUGUAACCAGCGGCACC-GCCGCUGUGCGAGAAGCUCCCCAGGUAAUUGC-GCUGCUGGUUGCCAUGGUGGCC-ACCGCCGCC-UCCGGU ---------------------(((((..(((((((..(((((((...-)))))))))))))).)))))((.(((((((((-...)).)))))))..((((((.-...))))))-...)). ( -46.00, z-score = -2.76, R) >droSim1.chr3R 13701018 95 - 27517382 ---------------------GGAGCUAUCUUGUAACCAGCGGCACC-GCCGCUGUGCGAGAAGCUCCCCAAGUAAUUGC-GCUGCUGGUUGCCAUGGUGGCC-ACCGCCGCC-UCCGGU ---------------------((((((.(((((((..(((((((...-))))))))))))))))))))(((.(((.....-..))))))..(((..((((((.-...))))))-...))) ( -45.40, z-score = -3.21, R) >droSec1.super_0 6733809 95 - 21120651 ---------------------GGAGCUAUCUUGUAUCCAGCGGCACC-GCCGCUGUGCGAGAAGCUCCCCAAGUAAUUGC-GCUGCUGGUUGCCAUGGUGGCC-ACCGCCGCC-UCCGGU ---------------------((((((.(((((((..(((((((...-))))))))))))))))))))(((.(((.....-..))))))..(((..((((((.-...))))))-...))) ( -45.40, z-score = -3.22, R) >droYak2.chr3R 11992453 95 - 28832112 ---------------------GGAGCAAUCUUGUAACCAGCGGCACC-GCCGCUGUGCGAGAAGCUCCCCAAGUAAUUGC-GCUGCUGGUUGCCAUGGUGGCC-ACCUCCGCC-UCCGGU ---------------------(((((..(((((((..(((((((...-)))))))))))))).)))))..........((-(..(.((((..(....)..)))-).)..))).-...... ( -41.50, z-score = -2.49, R) >droEre2.scaffold_4770 3708576 95 - 17746568 ---------------------GGAGCAAUCUUGUAACCAGCGGCACC-GCCGCUGUGCGAGAAGCUCCCCAAGUAAUUAC-GCUGCUGGUUGCCAUGGUGGCC-ACCUCCGCC-UCCGGU ---------------------(((((..(((((((..(((((((...-)))))))))))))).)))))(((.(((.....-..))))))..(((..(((((..-....)))))-...))) ( -39.40, z-score = -2.30, R) >droAna3.scaffold_13340 14060188 103 + 23697760 -------------AUGGCGGGGGUGCCACCUUGUAACCAGCGGCACC-GCCGCUGUGCGAGAAGCUGCCCAGAUAAUUGC-GUUGUUGGUUGCCGUAGUGGCC-GCCACCGCC-UCCAGU -------------..(((((.((((((((((((((..(((((((...-)))))))))))))..((.(((..((((((...-))))))))).))....)))).)-))).)))))-...... ( -47.00, z-score = -1.63, R) >dp4.chr2 5016025 95 - 30794189 ---------------------GGAGCUACCUUGUAGCCAGCGGCACC-GCCGCUGUGCGAGAAGCUACCCAAAUAGUUCC-GCUGCUGGUUGCCGUAGUGGCC-ACCGCCGCC-GCCAGU ---------------------(((((((....((((((((((((...-)))))))........))))).....)))))))-...((((((.((.(..((....-))..).)).-)))))) ( -39.40, z-score = -1.59, R) >droPer1.super_6 1145959 99 + 6141320 ---------------------GGAGCUACCUUGUAGCCAGCGGCACCCGCCGCUGUGCGAGAAGCUACCCAAAUAGUUCCCGCUGCUGGUUGCCGGAAUGGCCCACCGCCGCCCGCCAGU ---------------------(((((((....((((((((((((....)))))))........))))).....)))))))....((((((.(.(((...((....)).))).).)))))) ( -39.90, z-score = -1.89, R) >droWil1.scaffold_181089 5755780 116 - 12369635 UUUAGUGCUGCUGGUGGUGGUAGUGUUACCUUAUAGCCAGCUGCACC-GCCACUGUGGGAGAAGCUUCCCAAAUAGUUGC-GCUGCUGAUUGCCAUGAUGGCC-ACCGCCUCC-UCCAGU .........(((((.((.((..(((...((((((.((...(.(((.(-((.((((((((((....))))))..)))).))-).))).)...)).)))).)).)-))..)).))-.))))) ( -43.60, z-score = -1.59, R) >droVir3.scaffold_12855 5625957 94 + 10161210 ----------------------GCGCUAUUUUGUAGCCAGCGGCACC-GCCGCUGUGCGAGAAGCUGCCCAAAUAGUUGC-GCUGCUGAUUGCCAUAGUGACC-GCCACCGCC-GCCAGU ----------------------(.(((((...)))))).(((((...-((((((((((((..(((.((.(((....))).-)).)))..))).)))))))...-))....)))-)).... ( -32.20, z-score = -0.56, R) >droMoj3.scaffold_6540 5947134 90 + 34148556 --------------------------UAUCUUGUAGCCAGCGGCACC-GCCGCCGUGCGAGAAGCUACCCAAAUAAUUGC-GUUGCUGAUUGCCAUAAUGGCC-GCCGCCGCC-GCCAGU --------------------------.........((..(((((...-((.(((((((((..(((.((.(((....))).-)).)))..))))....))))).-)).))))).-)).... ( -29.80, z-score = -0.90, R) >droGri2.scaffold_14906 5791589 116 - 14172833 AACUGUGGUGGUGGUGGCUGUAGUGCACCCUUAUAGCCAGCGGCACC-GCCGCUGUGCGAGAAGCUGCCCAAAUAGUUGC-GCUGCUGAUUGCCAUAAUGACC-GCCACCACC-GCCAGU .....(((((((((((((.(((((((.........(((((((((...-))))))).))....(((((......)))))))-)))))((.....))........-)))))))))-)))).. ( -55.20, z-score = -4.17, R) >consensus _____________________GGAGCUAUCUUGUAGCCAGCGGCACC_GCCGCUGUGCGAGAAGCUCCCCAAAUAAUUGC_GCUGCUGGUUGCCAUAGUGGCC_ACCGCCGCC_UCCAGU .............................((((((..(((((((....))))))))))))).(((...................)))....(((.....))).................. (-18.98 = -19.41 + 0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:28 2011