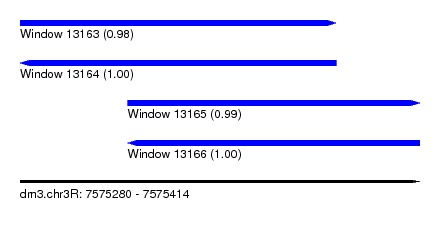
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,575,280 – 7,575,414 |
| Length | 134 |
| Max. P | 0.998401 |

| Location | 7,575,280 – 7,575,386 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 65.14 |
| Shannon entropy | 0.68529 |
| G+C content | 0.35539 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -11.23 |
| Energy contribution | -10.50 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
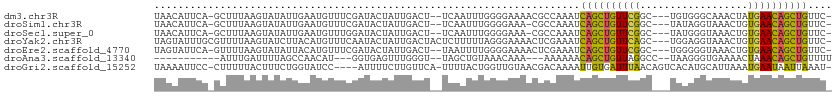
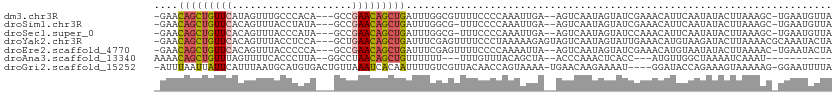
>dm3.chr3R 7575280 106 + 27905053 UAACAUUCA-GCUUUAAGUAUAUUGAAUGUUUCGAUACUAUUGACU--UCAAUUUGGGGAAAACGCCAAAUCAGCUGUUCGGC---UGUGGGCAAACUAUGAACAGCUGUUC- .((((((((-(..(.....)..)))))))))(((((...)))))..--...((((((.(....).))))))((((((((((..---.((......))..))))))))))...- ( -27.90, z-score = -2.34, R) >droSim1.chr3R 13697217 105 - 27517382 UAACAUUCA-GCUUUAAGUAUAUUGAAUGUUUCGAUACUAUUGACU--UCAAUUUGGGGAAA-CGCCAAAUCAGCUGUUCGGC---UAUAGGUAAACUGUGAACAGCUGUUC- .((((((((-(..(.....)..)))))))))(((((...)))))..--...(((((((....-).))))))(((((((((...---.((((.....)))))))))))))...- ( -28.80, z-score = -2.98, R) >droSec1.super_0 6730009 105 - 21120651 UAACAUUCA-GCUUUAAGUAUAUUGAAUGUUUGGAUACUAUUGACU--UCAAUUUGGGGAAA-CGCCAAAUCAGCUGUUCGGC---UAUGGGUAAACUGUGAACAGCUGUUC- .((((((((-(..(.....)..)))))))))...............--...(((((((....-).))))))(((((((((...---.((((.....)))))))))))))...- ( -27.30, z-score = -2.15, R) >droYak2.chr3R 11988670 109 - 28832112 UAGUAUUUGCGUUUUAAGUAUCUUACAUGUUUCAAUACUAUUGACUACUCUUUUUAGGGAAAACUCGAAAUCAGCUGUUCAGC---UGGAGGUAAACUGUGAACAGCUGUUC- (((((((.((((..((((...)))).))))...)))))))((((...((((....)))).....))))...((((((((((.(---.(........).)))))))))))...- ( -23.30, z-score = -0.64, R) >droEre2.scaffold_4770 3704857 106 - 17746568 UAGUAUUCA-GUUUUAAGUAUAUUACAUGUUUCGAUACUAUUGACU--UAAUUUUGGGGAAAACUCGAAAUCAGCUGUUCGGC---UGGGGGUAAACUGUGAACAGCUGUUC- ........(-(((...(((((.............)))))...))))--..((((((((.....))))))))((((((((((.(---.(........).)))))))))))...- ( -23.72, z-score = -1.22, R) >droAna3.scaffold_13340 14054944 92 + 23697760 -----------AUUUGAUUUUAGCCAACAU---GGUGAGUUUGGGU--UAGCUGUAAACAAA---AAAAAACAGCUGUUAGGCC--UAAGGGUGAAAACUAAACAGCUGUUUU -----------.((((.((.((((.(((..---((.....))..))--).)))).)).))))---..(((((((((((((((((--....))).....)).)))))))))))) ( -22.50, z-score = -0.98, R) >droGri2.scaffold_15252 12573421 106 - 17193109 UAAAAUUCC-CUUUUUACUUUCUGGUAUCC----AUUUUCUUGUUCA-UUUUACUGGUUGUAACGACAAAAUUGUGAUUUAACAGUCACAUGCAUUAAAUGAAUAAUUAAAU- .........-.....((((....))))...----((((..(((((((-(((...((((((...)))).....((((((......))))))..))..))))))))))..))))- ( -16.40, z-score = -1.14, R) >consensus UAACAUUCA_GCUUUAAGUAUAUUGAAUGUUUCGAUACUAUUGACU__UCAAUUUGGGGAAAACGCCAAAUCAGCUGUUCGGC___UAUGGGUAAACUGUGAACAGCUGUUC_ .......................................................................((((((((((..................)))))))))).... (-11.23 = -10.50 + -0.73)
| Location | 7,575,280 – 7,575,386 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 65.14 |
| Shannon entropy | 0.68529 |
| G+C content | 0.35539 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -10.00 |
| Energy contribution | -9.98 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7575280 106 - 27905053 -GAACAGCUGUUCAUAGUUUGCCCACA---GCCGAACAGCUGAUUUGGCGUUUUCCCCAAAUUGA--AGUCAAUAGUAUCGAAACAUUCAAUAUACUUAAAGC-UGAAUGUUA -((.(((((((((...(((.......)---)).)))))))))((((((.(....).))))))...--...........))..((((((((.............-)))))))). ( -23.22, z-score = -1.87, R) >droSim1.chr3R 13697217 105 + 27517382 -GAACAGCUGUUCACAGUUUACCUAUA---GCCGAACAGCUGAUUUGGCG-UUUCCCCAAAUUGA--AGUCAAUAGUAUCGAAACAUUCAAUAUACUUAAAGC-UGAAUGUUA -...(((((((((...(((.......)---)).)))))))))(((..(((-((((.....((((.--...))))......)))))......((....))..))-..))).... ( -23.60, z-score = -2.38, R) >droSec1.super_0 6730009 105 + 21120651 -GAACAGCUGUUCACAGUUUACCCAUA---GCCGAACAGCUGAUUUGGCG-UUUCCCCAAAUUGA--AGUCAAUAGUAUCCAAACAUUCAAUAUACUUAAAGC-UGAAUGUUA -...(((((((((...(((.......)---)).)))))))))((((((.(-....)))))))...--...............((((((((.............-)))))))). ( -23.32, z-score = -2.76, R) >droYak2.chr3R 11988670 109 + 28832112 -GAACAGCUGUUCACAGUUUACCUCCA---GCUGAACAGCUGAUUUCGAGUUUUCCCUAAAAAGAGUAGUCAAUAGUAUUGAAACAUGUAAGAUACUUAAAACGCAAAUACUA -(((((((((((((..(((.......)---))))))))))))..)))((.(.(((........))).).))..(((((((........((((...)))).......))))))) ( -21.26, z-score = -1.48, R) >droEre2.scaffold_4770 3704857 106 + 17746568 -GAACAGCUGUUCACAGUUUACCCCCA---GCCGAACAGCUGAUUUCGAGUUUUCCCCAAAAUUA--AGUCAAUAGUAUCGAAACAUGUAAUAUACUUAAAAC-UGAAUACUA -((((((((((((...(((.......)---)).)))))))))..))).(((.(((.......(((--(((..(((.(((........))).)))))))))...-.))).))). ( -16.90, z-score = -1.98, R) >droAna3.scaffold_13340 14054944 92 - 23697760 AAAACAGCUGUUUAGUUUUCACCCUUA--GGCCUAACAGCUGUUUUUU---UUUGUUUACAGCUA--ACCCAAACUCACC---AUGUUGGCUAAAAUCAAAU----------- ((((((((((((..((((........)--)))..))))))))))))..---((((.....(((((--((...........---..))))))).....)))).----------- ( -24.52, z-score = -3.94, R) >droGri2.scaffold_15252 12573421 106 + 17193109 -AUUUAAUUAUUCAUUUAAUGCAUGUGACUGUUAAAUCACAAUUUUGUCGUUACAACCAGUAAAA-UGAACAAGAAAAU----GGAUACCAGAAAGUAAAAAG-GGAAUUUUA -.......(((((((((......(((((........))))).((((((((((((.....)))..)-)).))))))))))----)))))...............-......... ( -14.90, z-score = 0.14, R) >consensus _GAACAGCUGUUCACAGUUUACCCAUA___GCCGAACAGCUGAUUUGGCGUUUUCCCCAAAAUGA__AGUCAAUAGUAUCGAAACAUUCAAUAUACUUAAAAC_UGAAUGUUA ....(((((((((....................)))))))))....................................................................... (-10.00 = -9.98 + -0.02)
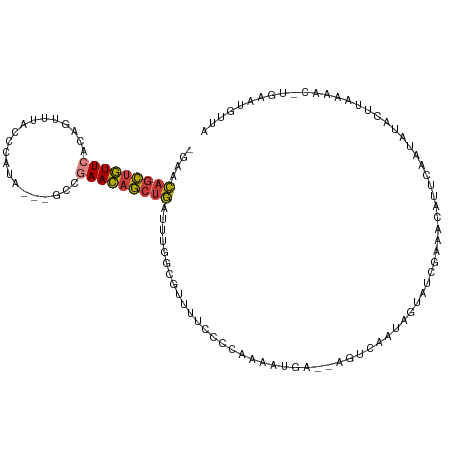
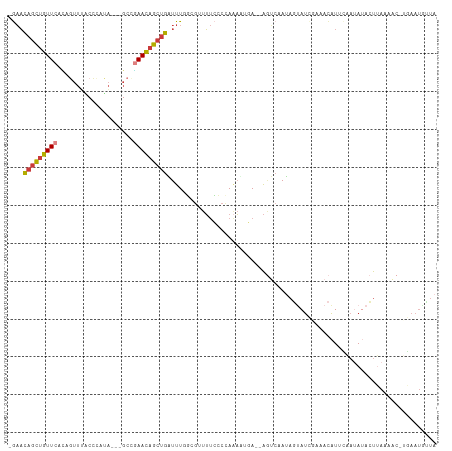
| Location | 7,575,316 – 7,575,414 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.92 |
| Shannon entropy | 0.65155 |
| G+C content | 0.35995 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -10.16 |
| Energy contribution | -9.96 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
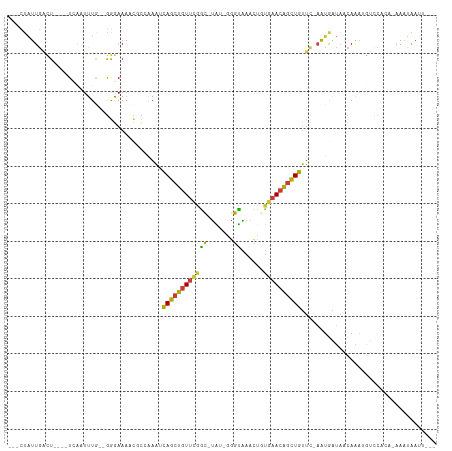
>dm3.chr3R 7575316 98 + 27905053 ---CUAUUGACU----UCAAUUUG--GGGAAAACGCCAAAUCAGCUGUUCGGC-UGU-GGGCAAACUAUGAACAGCUGUUC-AAUGAUAACAAAUGUCCCCA-AAAUAAUU--- ---.........----....((((--((((....(((....(((((....)))-)).-.)))......(((((....))))-).............))))))-))......--- ( -29.90, z-score = -3.68, R) >droSim1.chr3R 13697253 97 - 27517382 ---CUAUUGACU----UCAAUUUG--GGGAAA-CGCCAAAUCAGCUGUUCGGC-UAU-AGGUAAACUGUGAACAGCUGUUC-AAUGAUAACAAAUGUGCAAA-AAAUAAUU--- ---.((((((..----...(((((--((....-).))))))(((((((((...-.((-((.....))))))))))))).))-))))................-........--- ( -26.20, z-score = -3.40, R) >droSec1.super_0 6730045 97 - 21120651 ---CUAUUGACU----UCAAUUUG--GGGAAA-CGCCAAAUCAGCUGUUCGGC-UAU-GGGUAAACUGUGAACAGCUGUUC-AAUGAUAACAAAUGUCCCCA-AAAUAAUU--- ---.((((((..----...(((((--((....-).))))))(((((((((...-.((-((.....))))))))))))).))-))))................-........--- ( -24.90, z-score = -2.17, R) >droYak2.chr3R 11988707 99 - 28832112 ---CUAUUGACUA--CUCUUUUUA--GGGAAAACUCGAAAUCAGCUGUUCAGC-UGG-AGGUAAACUGUGAACAGCUGUUC-AAUGAUAACAAAAGGCCACA-AA-UAAUG--- ---.((((((...--((((....)--)))............((((((((((.(-.(.-.......).))))))))))).))-))))................-..-.....--- ( -20.90, z-score = -1.07, R) >droEre2.scaffold_4770 3704893 96 - 17746568 ---CUAUUGACU----UAAUUUUG--GGGAAAACUCGAAAUCAGCUGUUCGGC-UGG-GGGUAAACUGUGAACAGCUGUUC-AAUAAUAACAGA-GUCCCCA-AA-UAAUA--- ---.........----....((((--((((...(((.....((((((((((.(-.(.-.......).)))))))))))...-..........))-)))))))-))-.....--- ( -26.77, z-score = -2.55, R) >droAna3.scaffold_13340 14054970 95 + 23697760 ------UUGGGU----UAGCUGUA--AACAAA---AAAAAACAGCUGUUAGGCCUAA-GGGUGAAAACUAAACAGCUGUUUUAGUGAUAACAAAGUUUUGAGUAAACAAAA--- ------....((----(.(((..(--(((...---..(((((((((((((((((...-.))).....)).)))))))))))).((....))...))))..))).)))....--- ( -24.00, z-score = -2.37, R) >droGri2.scaffold_15252 12573453 114 - 17193109 UUCUUGUUCAUUUUACUGGUUGUAACGACAAAAUUGUGAUUUAACAGUCACAUGCAUUAAAUGAAUAAUUAAAUAUUGAACAUACAAACGCAUGCAAUGAAAUAAAUAAAUUGG ...((((((((((...((((((...)))).....((((((......))))))..))..)))))))))).....(((((..(((.(....).))))))))............... ( -15.30, z-score = 0.65, R) >consensus ___CUAUUGACU____UCAAUUUG__GGGAAAACGCCAAAUCAGCUGUUCGGC_UAU_GGGUAAACUGUGAACAGCUGUUC_AAUGAUAACAAAUGUCCACA_AAAUAAUU___ .........................................((((((((((.................)))))))))).................................... (-10.16 = -9.96 + -0.20)

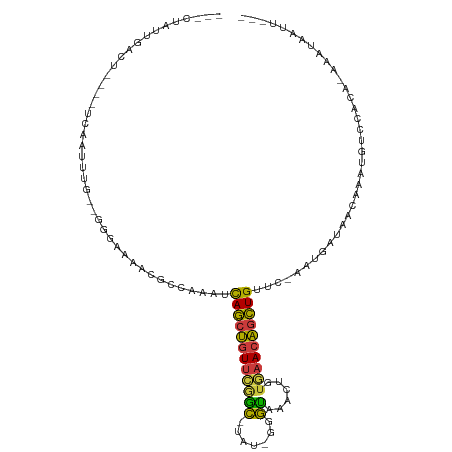
| Location | 7,575,316 – 7,575,414 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 64.92 |
| Shannon entropy | 0.65155 |
| G+C content | 0.35995 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -10.99 |
| Energy contribution | -10.48 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.998401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
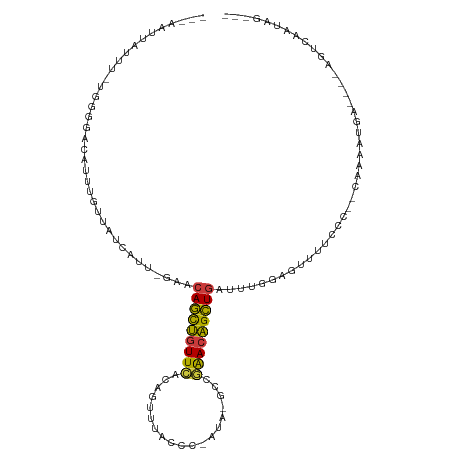
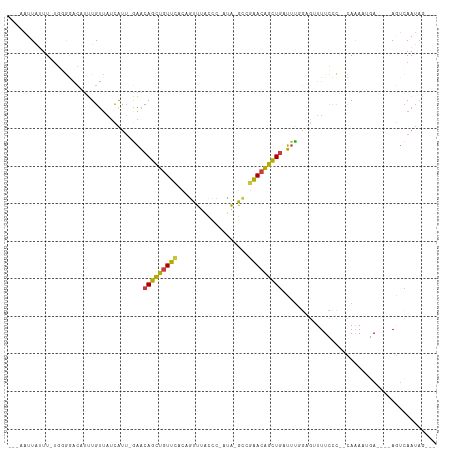
>dm3.chr3R 7575316 98 - 27905053 ---AAUUAUUU-UGGGGACAUUUGUUAUCAUU-GAACAGCUGUUCAUAGUUUGCCC-ACA-GCCGAACAGCUGAUUUGGCGUUUUCCC--CAAAUUGA----AGUCAAUAG--- ---..(((.((-((((((....(((((.....-((.(((((((((...(((.....-..)-)).))))))))).)))))))...))))--)))).)))----.........--- ( -27.00, z-score = -2.55, R) >droSim1.chr3R 13697253 97 + 27517382 ---AAUUAUUU-UUUGCACAUUUGUUAUCAUU-GAACAGCUGUUCACAGUUUACCU-AUA-GCCGAACAGCUGAUUUGGCG-UUUCCC--CAAAUUGA----AGUCAAUAG--- ---........-.................(((-((.(((((((((...(((.....-..)-)).)))))))))((((((.(-....))--)))))...----..)))))..--- ( -20.30, z-score = -2.20, R) >droSec1.super_0 6730045 97 + 21120651 ---AAUUAUUU-UGGGGACAUUUGUUAUCAUU-GAACAGCUGUUCACAGUUUACCC-AUA-GCCGAACAGCUGAUUUGGCG-UUUCCC--CAAAUUGA----AGUCAAUAG--- ---..(((.((-((((((....(((((.....-((.(((((((((...(((.....-..)-)).))))))))).)))))))-..))))--)))).)))----.........--- ( -27.40, z-score = -3.20, R) >droYak2.chr3R 11988707 99 + 28832112 ---CAUUA-UU-UGUGGCCUUUUGUUAUCAUU-GAACAGCUGUUCACAGUUUACCU-CCA-GCUGAACAGCUGAUUUCGAGUUUUCCC--UAAAAAGAG--UAGUCAAUAG--- ---.....-.(-((..(((((((.......((-(((((((((((((..(((.....-..)-))))))))))))..)))))........--..))))).)--)...)))...--- ( -21.23, z-score = -1.54, R) >droEre2.scaffold_4770 3704893 96 + 17746568 ---UAUUA-UU-UGGGGAC-UCUGUUAUUAUU-GAACAGCUGUUCACAGUUUACCC-CCA-GCCGAACAGCUGAUUUCGAGUUUUCCC--CAAAAUUA----AGUCAAUAG--- ---.....-((-(((((((-((..(((....)-)).(((((((((...(((.....-..)-)).))))))))).....)))...))))--))))....----.........--- ( -26.30, z-score = -3.83, R) >droAna3.scaffold_13340 14054970 95 - 23697760 ---UUUUGUUUACUCAAAACUUUGUUAUCACUAAAACAGCUGUUUAGUUUUCACCC-UUAGGCCUAACAGCUGUUUUUU---UUUGUU--UACAGCUA----ACCCAA------ ---(((((......)))))..((((...((..((((((((((((..((((......-..))))..))))))))))))..---..))..--.))))...----......------ ( -20.60, z-score = -3.17, R) >droGri2.scaffold_15252 12573453 114 + 17193109 CCAAUUUAUUUAUUUCAUUGCAUGCGUUUGUAUGUUCAAUAUUUAAUUAUUCAUUUAAUGCAUGUGACUGUUAAAUCACAAUUUUGUCGUUACAACCAGUAAAAUGAACAAGAA .......(((((...(((..((((((((...(((...((((......)))))))..))))))))..).)).))))).....((((((((((((.....)))..))).)))))). ( -18.60, z-score = -0.45, R) >consensus ___AAUUAUUU_UGGGGACAUUUGUUAUCAUU_GAACAGCUGUUCACAGUUUACCC_AUA_GCCGAACAGCUGAUUUGGAGUUUUCCC__CAAAAUGA____AGUCAAUAG___ ....................................(((((((((...................)))))))))......................................... (-10.99 = -10.48 + -0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:26 2011