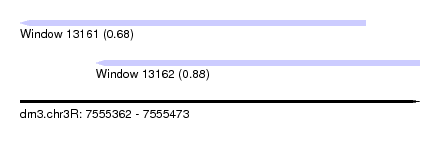
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,555,362 – 7,555,473 |
| Length | 111 |
| Max. P | 0.879737 |

| Location | 7,555,362 – 7,555,458 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.95 |
| Shannon entropy | 0.23155 |
| G+C content | 0.61066 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -28.36 |
| Energy contribution | -28.16 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.676662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
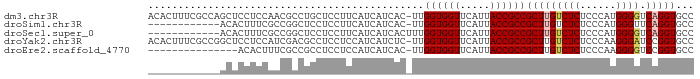
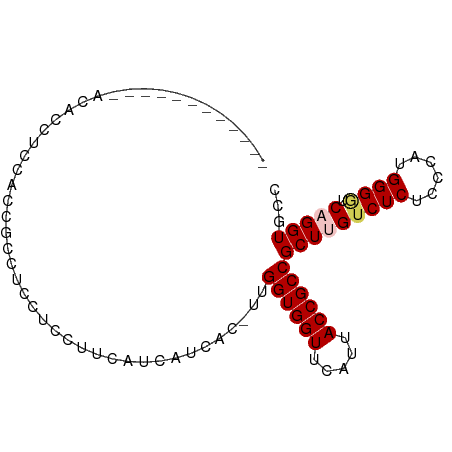
>dm3.chr3R 7555362 96 - 27905053 CCUCCAACGCCUGCUCCUUCAUCAUCAC-UUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAUGGGGUCAGGUGCCGGGGGACUGGCGCUCGGAUGG (((((...(((((((((........((.-.(((((((.....)))))))..))..........)))).)))))(((((....)))))....))).)) ( -39.87, z-score = -2.06, R) >droSim1.chr3R 13676564 84 + 27517382 ------------CCUCCUUCAUCAUCAC-UUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAUGGGUUCAGGUGCCGGGGGACUGGCGCUCGAAGGG ------------...(((((........-..((((((.....))))))(((((.(((......)))..)))))(((((....)))))....))))). ( -35.20, z-score = -2.37, R) >droSec1.super_0 6710030 85 + 21120651 ------------CCUCCUUCAUCAUCACUUUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAUGGGGUCAGGUGCCGGGGGACUGGCGCUCGGAUGG ------------(((((..............((((((.....))))))(((((.((((.....)))).)))))(((((....)))))....))).)) ( -33.70, z-score = -1.46, R) >droYak2.chr3R 11968539 88 + 28832112 --------GACGCCUCCUCCAUCAUCUC-UUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAAGGGAUCCGGUGCCGGGGGACUGGCAUUUGGUUGG --------...(((((((((..((((..-..((((((.....))))))....((((((....))))))..))))..))))))..))).......... ( -31.90, z-score = -0.71, R) >droEre2.scaffold_4770 3685006 84 + 17746568 ------------CCUCCUCCAUCAUCAC-UUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAAGGGGUCCGGUGCCGGGGGAUUGGCAUUCGGAUGG ------------......(((((.....-..((((((.....))))))(((.((((((((....))).(((....)))))))).))).....))))) ( -32.20, z-score = -0.89, R) >consensus ____________CCUCCUUCAUCAUCAC_UUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAUGGGGUCAGGUGCCGGGGGACUGGCGCUCGGAUGG ..................(((((........((((((.....)))))).........(((....))).(..(((((((....)))))))..)))))) (-28.36 = -28.16 + -0.20)

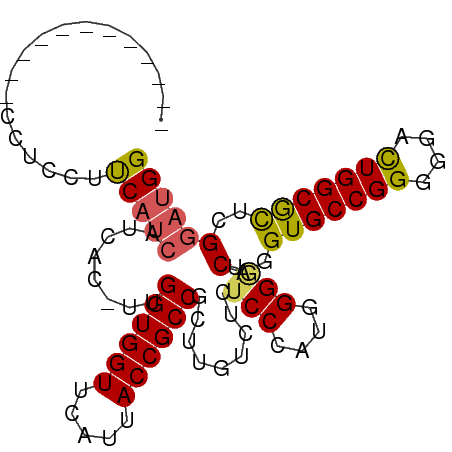
| Location | 7,555,383 – 7,555,473 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 78.80 |
| Shannon entropy | 0.35067 |
| G+C content | 0.58230 |
| Mean single sequence MFE | -23.21 |
| Consensus MFE | -15.44 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7555383 90 - 27905053 ACACUUUCGCCAGCUCCUCCAACGCCUGCUCCUUCAUCAUCAC-UUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAUGGGGUCAGGUGCC ......................((((((((((........((.-.(((((((.....)))))))..))..........)))).)))))).. ( -25.27, z-score = -2.04, R) >droSim1.chr3R 13676585 78 + 27517382 ------------ACACUUUCGCCGGCUCCUCCUUCAUCAUCAC-UUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAUGGGUUCAGGUGCC ------------...........(((.................-..((((((.....))))))(((((.(((......)))..)))))))) ( -20.00, z-score = -1.44, R) >droSec1.super_0 6710051 79 + 21120651 ------------ACACUUUCGCCGGCUCCUCCUUCAUCAUCACUUUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAUGGGGUCAGGUGCC ------------.......((((((((((...........((...(((((((.....)))))))..))..........)))))).)))).. ( -22.50, z-score = -1.74, R) >droYak2.chr3R 11968560 90 + 28832112 ACACUUUCGCCGGCUCCUCCAUCGACGCCUCCUCCAUCAUCUC-UUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAAGGGAUCCGGUGCC .......((((((.(((..........................-..((((((.....)))))).((((.......))))))).)))))).. ( -25.90, z-score = -2.16, R) >droEre2.scaffold_4770 3685027 75 + 17746568 ---------------ACACUUUCGCCGCCUCCUCCAUCAUCAC-UUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAAGGGGUCCGGUGCC ---------------.......(((((.(((((.......((.-.(((((((.....)))))))..)).........)))))..))))).. ( -22.39, z-score = -2.12, R) >consensus ____________ACACCUCCACCGCCUCCUCCUUCAUCAUCAC_UUGGUGGUUCAUUACCGCCGCUUGUCUCUCCCAUGGGGUCAGGUGCC ..............................................((((((.....))))))(((((((((......)))).)))))... (-15.44 = -15.92 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:23 2011