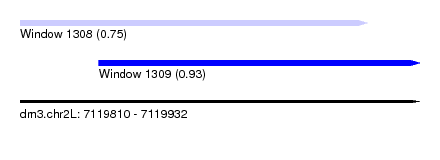
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,119,810 – 7,119,932 |
| Length | 122 |
| Max. P | 0.934602 |

| Location | 7,119,810 – 7,119,916 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.27 |
| Shannon entropy | 0.42510 |
| G+C content | 0.48611 |
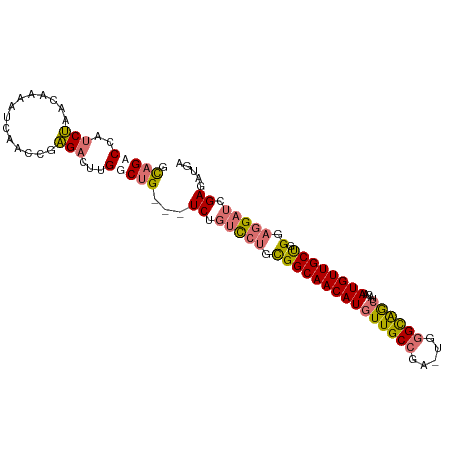
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -18.15 |
| Energy contribution | -18.34 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7119810 106 + 23011544 UCAUCGGUCGAAGUACCAGU-CAAAGCAGACCAUCUAACAAAAUCAACCGAGACUUGUCUG---UCUGUCCUGCGGCAACAUGUUGCCGA-UGGGCAGCUACAAUGUUGCU .(((((((......)))...-....((((..((.(..((((..((....))...))))..)---..))..))))(((((....)))))))-))((((((......)))))) ( -30.20, z-score = -1.26, R) >droSim1.chr2L 6908977 106 + 22036055 UCAUCGGUCGAACUACCAGC-CAAAGCAGACCAUCUAACAAAAUCAACGGAGACUUGGCUG---UCUGUCCUGCGGCAACAUGUUGCCGA-UGGGCAGCUACAAUGUUGCU .....(((((......).((-....)).))))............(((((....).((((((---((((((....(((((....)))))))-))))))))))....)))).. ( -32.60, z-score = -1.43, R) >droSec1.super_3 2643539 106 + 7220098 UCAUCGGUCGAACUACCAGC-CAAAGCAGACCAUCUAACAAAAUCAACCGAGACUUGUCUG---UCUGUCCUGCGGCAACAUGUUGCCGA-UGGGCAGCUACAAUGUUGCU ...(((((.((.......((-....))((.....)).......)).)))))(((((((..(---.((((((..((((((....)))))).-.))))))).)))).)))... ( -31.10, z-score = -1.73, R) >droWil1.scaffold_180772 3183397 99 - 8906247 ------------UCAUCAAUACUAAAUAGACCCCCAGACACAAUCGAAACUGUUUUGGCCGGCAUCUUGUUGUUGGCAACAUGUUGCCAACUGAGUGAGAUGAAUGUUGCC ------------....(((((.........((.(((((.(((........))))))))..))(((((..((((((((((....))))))))..))..)))))..))))).. ( -26.50, z-score = -1.68, R) >consensus UCAUCGGUCGAACUACCAGC_CAAAGCAGACCAUCUAACAAAAUCAACCGAGACUUGGCUG___UCUGUCCUGCGGCAACAUGUUGCCGA_UGGGCAGCUACAAUGUUGCU ...............(((.......(((((........(((..((....))...))).......)))))....((((((....))))))..)))(((((......))))). (-18.15 = -18.34 + 0.19)

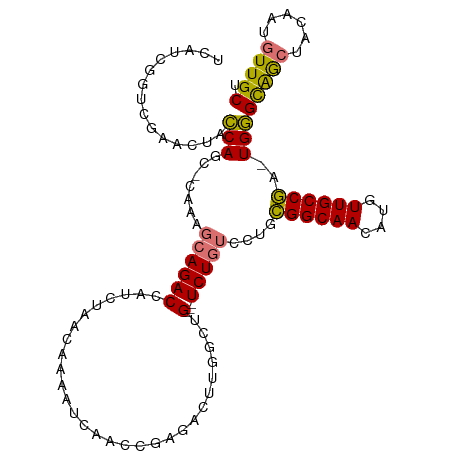
| Location | 7,119,834 – 7,119,932 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.50 |
| Shannon entropy | 0.39038 |
| G+C content | 0.50760 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -21.35 |
| Energy contribution | -22.16 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7119834 98 + 23011544 GCAGACCAUCUAACAAAAUCAACCGAGACUUGUCUG---UCUGUCCUGCGGCAACAUGUUGCCGA-UGGGCAGCUACAAUGUUGCUGGGAGGAUCGAGAUGA ......(((((..............((((......)---)))(((((.(((((((((((((((..-..))))))....)))))))).).)))))..))))). ( -34.30, z-score = -2.15, R) >droSim1.chr2L 6909001 98 + 22036055 GCAGACCAUCUAACAAAAUCAACGGAGACUUGGCUG---UCUGUCCUGCGGCAACAUGUUGCCGA-UGGGCAGCUACAAUGUUGCUGGGAGGAUCGAGAUGA ......(((((..............((((......)---)))(((((.(((((((((((((((..-..))))))....)))))))).).)))))..))))). ( -34.30, z-score = -1.96, R) >droSec1.super_3 2643563 98 + 7220098 GCAGACCAUCUAACAAAAUCAACCGAGACUUGUCUG---UCUGUCCUGCGGCAACAUGUUGCCGA-UGGGCAGCUACAAUGUUGCUGGGAGGAUCGAGAUGA ......(((((..............((((......)---)))(((((.(((((((((((((((..-..))))))....)))))))).).)))))..))))). ( -34.30, z-score = -2.15, R) >droWil1.scaffold_180772 3183410 96 - 8906247 AUAGACCCCCAGACACAAUCGAAACUGUUUUGGCCGGCAUCUUGUUGUUGGCAACAUGUUGCCAACUGAGUGAGAUGAAUGUUGCCCGCGUG-UUGA----- ...........(((((...((((.....))))((.(((((((..((((((((((....))))))))..))..))).......)))).)))))-))..----- ( -28.41, z-score = -1.57, R) >consensus GCAGACCAUCUAACAAAAUCAACCGAGACUUGGCUG___UCUGUCCUGCGGCAACAUGUUGCCGA_UGGGCAGCUACAAUGUUGCUGGGAGGAUCGAGAUGA .(((((..(((..............)))...)))))...((.(((((.(((((((((((((((.....))))))....)))))))).).))))).))..... (-21.35 = -22.16 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:55 2011