
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,485,928 – 7,486,079 |
| Length | 151 |
| Max. P | 0.823936 |

| Location | 7,485,928 – 7,486,039 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 85.60 |
| Shannon entropy | 0.26860 |
| G+C content | 0.41909 |
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -25.37 |
| Energy contribution | -24.74 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
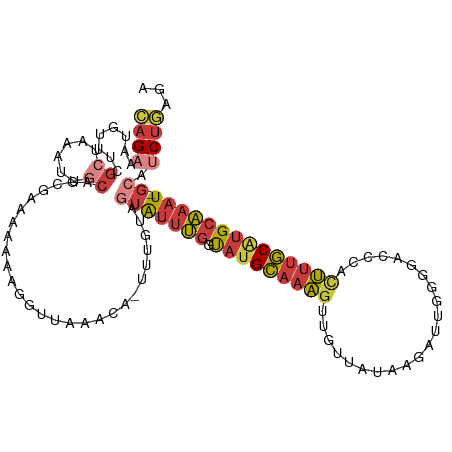
>dm3.chr3R 7485928 111 - 27905053 CA-UUUGCAGUAUUUGGUAUGCAAAGUUGCUAUAAGAUUGGGGACCCACUUUGCGUGCAAAUGCAUCUGAGAAAUGAUGCUCGAGGCAUUCAAAUGAUGAAAUAUGGCACCA ..-((((((..........))))))(.(((((((....(((....))).((((.((((....(((((........))))).....)))).))))........))))))).). ( -29.50, z-score = -0.53, R) >droSim1.chr3R 13607221 108 + 27517382 CA-UUUGCAGUAUUUGGCAUGCAAAGUGGUUAUAAGAUUGGGGACCCACUUUGCAUGCAAAUGCAUCUGAGA---GAUGCUCGAGGCAUUCAAAUGAUGAAAUAUGGCACCA ..-..(((.((((((.(((((((((((((..........(....)))))))))))))).....((((.....---(((((.....))))).....)))))))))).)))... ( -37.00, z-score = -2.78, R) >droSec1.super_0 6641082 108 + 21120651 CA-UUUGCAGUAUUUGGCAUGCAAAGUGGUUAUAAGAUUGGGGACCCACUUUGCAUGCAAAUGCAUCUGAGA---GAUGCUCGAGGCAUUCAAAUGAUGAAAUAUGGCACCA ..-..(((.((((((.(((((((((((((..........(....)))))))))))))).....((((.....---(((((.....))))).....)))))))))).)))... ( -37.00, z-score = -2.78, R) >droYak2.chr3R 11895844 107 + 28832112 CA-UUUGUAGUGUUUGGUAUGCAAAGUUGUUAUAAGAUU-GGAACCCUAUUUGCAUGCAAAUGCAUCUGAGA---GAUGCUCGUGGCAUCCGAAUGAUGAAAUAUGGCACUA ..-....(((((((..(((((((((...(((........-..)))....))))))))).....((((...(.---(((((.....))))))....))))......))))))) ( -26.90, z-score = -0.69, R) >droEre2.scaffold_4770 3613584 107 + 17746568 CA-UUUGUCGUAUUUGGUAUGCAAAGUUGUUAUAAGA-UGGGGACCCUCUUUGCAUGCAAAUGCAUCUGAAA---GAUGCUCGAGGCAUCCGAAUGAUGAAAUAUGGCACCA ..-..((((((((((.((((((((((...........-.((....)).)))))))))).....((((.....---(((((.....))))).....))))))))))))))... ( -33.62, z-score = -2.27, R) >droAna3.scaffold_13340 13948275 108 - 23697760 CAUUUUGUGGUGUUUGGUAUGCAAAGCAGUUGGGUCGAU-UGGACCUCUUUUGCAUGCAAAUGGACCUCAGA---GAUGCCCCAGGCAUUUAAAUGAUGAAAUAUGGCAAUC ....((((.((((((.((((((((((.....(((((...-..))))).)))))))))).......(.(((.(---((((((...)))))))...))).))))))).)))).. ( -33.90, z-score = -1.67, R) >consensus CA_UUUGCAGUAUUUGGUAUGCAAAGUUGUUAUAAGAUUGGGGACCCACUUUGCAUGCAAAUGCAUCUGAGA___GAUGCUCGAGGCAUUCAAAUGAUGAAAUAUGGCACCA .....(((.((((((.((((((((((..(((...........)))...)))))))))).....((((........(((((.....))))).....)))))))))).)))... (-25.37 = -24.74 + -0.63)



| Location | 7,485,968 – 7,486,079 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.09 |
| Shannon entropy | 0.51678 |
| G+C content | 0.39094 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -11.52 |
| Energy contribution | -11.97 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.564769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7485968 111 - 27905053 CAGAAAUGCUUCACUAAAUUGCAGCGAGAAAAAGGUUAAACA-UUUGCAGUAUUUGGUAUGCAAAGUUGCUAUAAGAUUGGGGACCCACUUUGCGUGCAAAU-GCAUCUGAGA ((((..((((.((......)).))))................-......(((((((.(((((((((((.(((......))).)....)))))))))))))))-)).))))... ( -26.70, z-score = -0.39, R) >droSim1.chr3R 13607258 111 + 27517382 CAGAAAUGUUUCGCUAAAUUGCAGCGAGAAAAAGGUUAAACA-UUUGCAGUAUUUGGCAUGCAAAGUGGUUAUAAGAUUGGGGACCCACUUUGCAUGCAAAU-GCAUCUGAGA ((((....(((((((.......))))))).............-......(((((((.((((((((((((..........(....))))))))))))))))))-)).))))... ( -38.00, z-score = -4.01, R) >droSec1.super_0 6641119 111 + 21120651 CAGAAAUGUUUCGCUAAAUUGCAGCGAGAAAAAGGUUAAACA-UUUGCAGUAUUUGGCAUGCAAAGUGGUUAUAAGAUUGGGGACCCACUUUGCAUGCAAAU-GCAUCUGAGA ((((....(((((((.......))))))).............-......(((((((.((((((((((((..........(....))))))))))))))))))-)).))))... ( -38.00, z-score = -4.01, R) >droYak2.chr3R 11895881 110 + 28832112 CAGAAACGUUUCGCUAAAUUGCAGCAAAAAAAAGGUUAAACA-UUUGUAGUGUUUGGUAUGCAAAGUUGUUAUAAGAUU-GGAACCCUAUUUGCAUGCAAAU-GCAUCUGAGA ((((........((......)).(((..........((((((-(.....)))))))(((((((((...(((........-..)))....)))))))))...)-)).))))... ( -23.70, z-score = -0.49, R) >droEre2.scaffold_4770 3613621 108 + 17746568 CAGAAACGUUUCGCUAAAUUGCAGC--AAAAAAGGUUAAACA-UUUGUCGUAUUUGGUAUGCAAAGUUGUUAUAAGA-UGGGGACCCUCUUUGCAUGCAAAU-GCAUCUGAAA (((((((.(((.(((.......)))--...))).))).....-......(((((((.(((((((((...........-.((....)).))))))))))))))-)).))))... ( -25.62, z-score = -1.04, R) >droAna3.scaffold_13340 13948312 111 - 23697760 GAGAAAUCAUUCGCCAAAUUACUUAAGAAAAAAGGUUUGACAUUUUGUGGUGUUUGGUAUGCAAAGCAGUUGGGUCGAU-UGGACCUCUUUUGCAUGCAAAU-GGACCUCAGA (((....((..(((((......(((((........))))).......)))))..))((((((((((.....(((((...-..))))).))))))))))....-....)))... ( -27.62, z-score = -0.63, R) >droPer1.super_3 3381414 108 - 7375914 UCGAGUUCAUUCGCCAAAUUACAG---AAAAAAUACUAAACAAAUAUUAAGGUAGCAUCUGU-AUGCAAAUGUGGCACC-GGCACACUUGCCACUUGCCACUCGCAGCCCGAA (((.((.............(((((---(.....((((.............))))...)))))-)(((....((((((..-((((....))))...))))))..))))).))). ( -26.02, z-score = -1.89, R) >consensus CAGAAAUGUUUCGCUAAAUUGCAGCGAAAAAAAGGUUAAACA_UUUGUAGUAUUUGGUAUGCAAAGUUGUUAUAAGAUUGGGGACCCACUUUGCAUGCAAAU_GCAUCUGAGA ((((.............(((((((....................))))))).....((((((((((......................))))))))))........))))... (-11.52 = -11.97 + 0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:18 2011