| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,475,685 – 7,475,789 |
| Length | 104 |
| Max. P | 0.967290 |

| Location | 7,475,685 – 7,475,789 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.98 |
| Shannon entropy | 0.52935 |
| G+C content | 0.51089 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -17.71 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
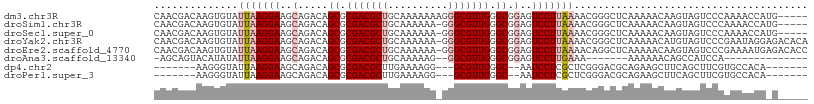
>dm3.chr3R 7475685 104 + 27905053 CAACGACAAGUGUAUUAAGGAAGCAGACAGCGCGACGCUGCAAAAAAAGGGCGUUGGGCGGAGUCCUUAAAACGGGCUCAAAAACAAGUAGUCCCAAAACCAUG----- ...........((.(((((((..(.....((.(((((((..........))))))).)).)..))))))).))((((((........).)))))..........----- ( -28.50, z-score = -1.52, R) >droSim1.chr3R 23708907 103 - 27517382 CAACGACAAGUGUAUUAAGGAAGCAGACAGCGCGACGCUGCAAAAAA-GGGCGUUGGGCGGAGUCCUUAAAACGGGCUCAAAAACAAGUAGUCCCAAAACCAUG----- ...........((.(((((((..(.....((.(((((((........-.))))))).)).)..))))))).))((((((........).)))))..........----- ( -28.60, z-score = -1.57, R) >droSec1.super_0 6630913 103 - 21120651 CAACGACAAGUGUAUUAAGGAAGCAGACAGCGCGACGCUGCAAAAAA-GGGCGUUGGGCGGAGUCCUUAAAACGGGCUCAAAAACAAGUAGUCCCAAAACCAUG----- ...........((.(((((((..(.....((.(((((((........-.))))))).)).)..))))))).))((((((........).)))))..........----- ( -28.60, z-score = -1.57, R) >droYak2.chr3R 11885517 108 - 28832112 CAACGACAAGUGUAUUAAGGAAGCAGACAGCGCGACGCUGCAAAAAA-GGGCGUUGGGCGGAGUCCUUAAAACGGGCUCAAAAACAUGUAGUCCCGAAUAGGAGACACA .........((((.(((((((..(.....((.(((((((........-.))))))).)).)..)))))))..((((((((......)).)).))))........)))). ( -34.30, z-score = -2.95, R) >droEre2.scaffold_4770 3603116 108 - 17746568 CAACGACAAGUGUAUUAAGGAAGCAGACAGCGCGACGCUGCAAAAAA-GGGCGUUGGGCGGAGUCCUUAAAACAGGCUCAAAAACAAGUAGUCCCGAAAAUGAGACACC ....(((...(((.(((((((..(.....((.(((((((........-.))))))).)).)..))))))).))).(((........))).)))................ ( -27.30, z-score = -1.19, R) >droAna3.scaffold_13340 13937117 85 + 23697760 -AGCAGUACAUAUAUUAAGGAAGCAGACAGCGCGACGCUGCAAAAAG--GGCGUUGGGCGGAGUCCUUGAAA-------AAAAAACAGCCAUCCA-------------- -.((.((.......(((((((..(.....((.(((((((........--))))))).)).)..)))))))..-------.....)).))......-------------- ( -23.54, z-score = -1.51, R) >dp4.chr2 20619875 90 + 30794189 -------AAGGGUAUUAAGGAAGCAGACAGCGCGACGCUUGAAAAGG---GCGUUGGGC--AAUCCUCGCUCGGGACGCAGAAGCUUCAGCUUCGUGCCACA------- -------...(((((.......((.....((.((((((((.....))---)))))).))--..((((.....)))).)).(((((....))))))))))...------- ( -32.80, z-score = -1.37, R) >droPer1.super_3 3371117 90 + 7375914 -------AAGGGUAUUAAGGAAGCAGACAGCGCGACGCUUGAAAAGG---GCGUUGGGC--AAUCCUCGCUCGGGACGCAGAAGCUUCAGCUUCGUGCCACA------- -------...(((((.......((.....((.((((((((.....))---)))))).))--..((((.....)))).)).(((((....))))))))))...------- ( -32.80, z-score = -1.37, R) >consensus CAACGACAAGUGUAUUAAGGAAGCAGACAGCGCGACGCUGCAAAAAA_GGGCGUUGGGCGGAGUCCUUAAAACGGGCUCAAAAACAAGUAGUCCCAAAAACA_G_____ ..............(((((((..(.....((.(((((((..........))))))).)).)..)))))))....................................... (-17.71 = -18.48 + 0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:14 2011