| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,118,349 – 7,118,404 |
| Length | 55 |
| Max. P | 0.972110 |

| Location | 7,118,349 – 7,118,404 |
|---|---|
| Length | 55 |
| Sequences | 9 |
| Columns | 55 |
| Reading direction | forward |
| Mean pairwise identity | 89.63 |
| Shannon entropy | 0.21648 |
| G+C content | 0.44913 |
| Mean single sequence MFE | -18.21 |
| Consensus MFE | -11.44 |
| Energy contribution | -12.13 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
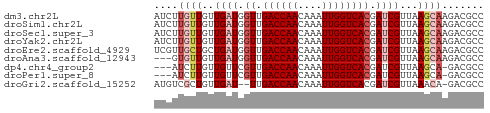
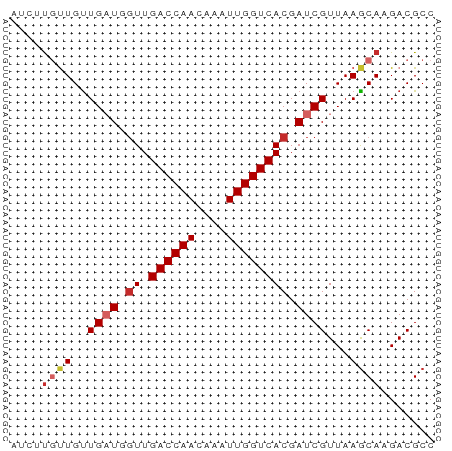
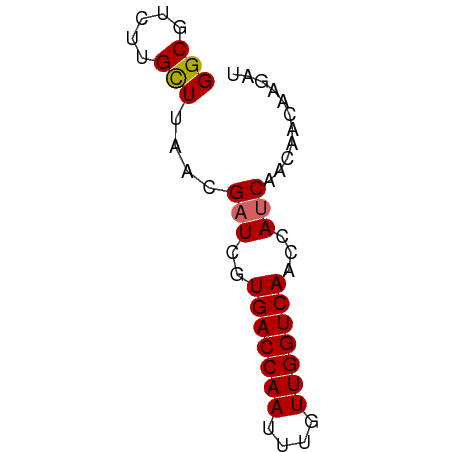
>dm3.chr2L 7118349 55 + 23011544 AUCUUGUUGUUGAUGGUUGACCAACAAAUUGGUCACGAUCGUUAAGCAAGACGCC .(((((((..((((.((.((((((....)))))))).))))...))))))).... ( -20.80, z-score = -4.01, R) >droSim1.chr2L 6907485 55 + 22036055 AUCUUGUUGUUGAUGGUUGACCAACAAAUUGGUCACGAUCGUUAAGCAAGACGCC .(((((((..((((.((.((((((....)))))))).))))...))))))).... ( -20.80, z-score = -4.01, R) >droSec1.super_3 2642185 55 + 7220098 AUCUUGUUGUUGAUGGUUGACCAACAAAUUGGUCACGAUCGUUAAGCAAGACGCC .(((((((..((((.((.((((((....)))))))).))))...))))))).... ( -20.80, z-score = -4.01, R) >droYak2.chr2L 16536133 55 - 22324452 AUCUUGUUGUUGAUGGUUGACCAACAAAUUGGUCACGAUCGUUAAGCAAGACGCC .(((((((..((((.((.((((((....)))))))).))))...))))))).... ( -20.80, z-score = -4.01, R) >droEre2.scaffold_4929 16033856 55 + 26641161 UCGUUGCUGCUGAUGGUUGACCAACAAAUUGGUCACGAUCGUUAAGCAAGACGCC .((((..(((((((((((((((((....)))))))..)))))).)))).)))).. ( -19.80, z-score = -3.14, R) >droAna3.scaffold_12943 969226 52 - 5039921 ---GUGUUGUUGAUGGUUGACCAACAAAUUGGUCACGAUCGUUAAGCAAGACGCC ---((.((((((((((((((((((....)))))))..)))))).))))).))... ( -18.30, z-score = -3.09, R) >dp4.chr4_group2 742637 51 + 1235136 ---AUCUUGUUGUUCGUUGACCAACAAAUUGGUCACGAUCGUUAAGCA-GACGCC ---..((((.((.((((.((((((....)))))))))).)).))))..-...... ( -13.90, z-score = -1.97, R) >droPer1.super_8 811788 51 - 3966273 ---AUCUUGUUGUUCGUUGACCAACAAAUUGGUCACGAUCGUUAAGCA-GACGCC ---..((((.((.((((.((((((....)))))))))).)).))))..-...... ( -13.90, z-score = -1.97, R) >droGri2.scaffold_15252 1434837 52 - 17193109 AUGUCGCUGUUGAU--UUGACCAACAAAUUGGUCACGAUCGUUAAACA-GACGCC ....((((((((((--((((((((....))))))).))))....))))-).)).. ( -14.80, z-score = -2.24, R) >consensus AUCUUGUUGUUGAUGGUUGACCAACAAAUUGGUCACGAUCGUUAAGCAAGACGCC ....((((..((((.((.((((((....)))))))).))))...))))....... (-11.44 = -12.13 + 0.69)

| Location | 7,118,349 – 7,118,404 |
|---|---|
| Length | 55 |
| Sequences | 9 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 89.63 |
| Shannon entropy | 0.21648 |
| G+C content | 0.44913 |
| Mean single sequence MFE | -13.89 |
| Consensus MFE | -10.40 |
| Energy contribution | -10.52 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.607905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7118349 55 - 23011544 GGCGUCUUGCUUAACGAUCGUGACCAAUUUGUUGGUCAACCAUCAACAACAAGAU ...((((((......(((..(((((((....)))))))...))).....)))))) ( -14.50, z-score = -2.15, R) >droSim1.chr2L 6907485 55 - 22036055 GGCGUCUUGCUUAACGAUCGUGACCAAUUUGUUGGUCAACCAUCAACAACAAGAU ...((((((......(((..(((((((....)))))))...))).....)))))) ( -14.50, z-score = -2.15, R) >droSec1.super_3 2642185 55 - 7220098 GGCGUCUUGCUUAACGAUCGUGACCAAUUUGUUGGUCAACCAUCAACAACAAGAU ...((((((......(((..(((((((....)))))))...))).....)))))) ( -14.50, z-score = -2.15, R) >droYak2.chr2L 16536133 55 + 22324452 GGCGUCUUGCUUAACGAUCGUGACCAAUUUGUUGGUCAACCAUCAACAACAAGAU ...((((((......(((..(((((((....)))))))...))).....)))))) ( -14.50, z-score = -2.15, R) >droEre2.scaffold_4929 16033856 55 - 26641161 GGCGUCUUGCUUAACGAUCGUGACCAAUUUGUUGGUCAACCAUCAGCAGCAACGA ..(((.(((((....(((..(((((((....)))))))...))))))))..))). ( -15.20, z-score = -1.45, R) >droAna3.scaffold_12943 969226 52 + 5039921 GGCGUCUUGCUUAACGAUCGUGACCAAUUUGUUGGUCAACCAUCAACAACAC--- (((.....)))....(((..(((((((....)))))))...)))........--- ( -11.50, z-score = -1.22, R) >dp4.chr4_group2 742637 51 - 1235136 GGCGUC-UGCUUAACGAUCGUGACCAAUUUGUUGGUCAACGAACAACAAGAU--- (((...-.)))....(.((((((((((....)))))).)))).)........--- ( -12.70, z-score = -1.37, R) >droPer1.super_8 811788 51 + 3966273 GGCGUC-UGCUUAACGAUCGUGACCAAUUUGUUGGUCAACGAACAACAAGAU--- (((...-.)))....(.((((((((((....)))))).)))).)........--- ( -12.70, z-score = -1.37, R) >droGri2.scaffold_15252 1434837 52 + 17193109 GGCGUC-UGUUUAACGAUCGUGACCAAUUUGUUGGUCAA--AUCAACAGCGACAU ..((.(-((((....(((..(((((((....))))))).--)))))))))).... ( -14.90, z-score = -2.04, R) >consensus GGCGUCUUGCUUAACGAUCGUGACCAAUUUGUUGGUCAACCAUCAACAACAAGAU (((.....)))....(((..(((((((....)))))))...)))........... (-10.40 = -10.52 + 0.12)

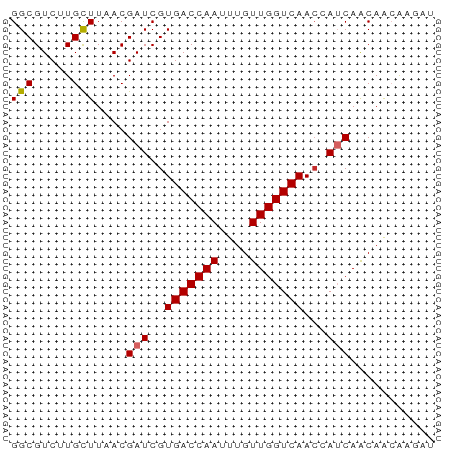
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:54 2011