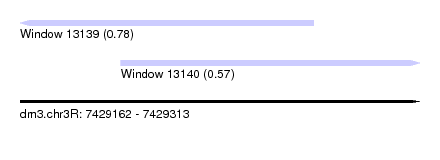
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,429,162 – 7,429,313 |
| Length | 151 |
| Max. P | 0.777721 |

| Location | 7,429,162 – 7,429,273 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.37 |
| Shannon entropy | 0.26683 |
| G+C content | 0.43889 |
| Mean single sequence MFE | -28.61 |
| Consensus MFE | -21.14 |
| Energy contribution | -22.67 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7429162 111 - 27905053 UUACGUGC----CUACUGCACAUUCUGUUAUUGUUUGCCA---UCGCUCGAGUGAACUUUGAUUGGCAAACAUUUCGGAAGGCAGCAGCCUCA--UCGUGAUCUGCAUCAACAUCUCCAU (((((((.----((.((((...(((((....(((((((((---(((...(((....)))))).)))))))))...))))).)))).))...))--.)))))................... ( -32.10, z-score = -2.09, R) >droAna3.scaffold_13340 13874020 104 - 23697760 UCACAUGCCAUUCCAUUAGACAUU-AAUUAUUGUUUGCCACCGUCGCUCGAGUGAACUUUGAUUGGCAAACAUUUCCGAAG---GCAGUGUAGCUUCAACAUCUUCAU------------ .(((.((((.(((.(((((...))-)))...(((((((((..((((...(((....)))))))))))))))).....))))---))))))..................------------ ( -23.50, z-score = -1.59, R) >droEre2.scaffold_4770 3555355 108 + 17746568 UUACGUGC----CUACUGCAUAUUUUGUUAUUGUUUGCCA---CCGCUCGAGUGAACUUUGAUUGGCAAACAUUUCGGAAG---GCAGCCUCA--UCGUGAUCUGCAUCAACAUCUUCAU (((((...----...((((...(((((....(((((((((---...(..(((....))).)..)))))))))...))))).---)))).....--.)))))................... ( -27.30, z-score = -1.30, R) >droYak2.chr3R 11838160 110 + 28832112 UUACGUGC----CUACUGCAUAUUUUGUUAUUGUUUGCCA---CCGCUCGAGUGAACUUUGAUUGGCAAACAUUUCGGAAG---GCAGCCUAAUCUCGUGAUCUGCAUCAACAUCUCCAU (((((...----...((((...(((((....(((((((((---...(..(((....))).)..)))))))))...))))).---))))........)))))................... ( -26.14, z-score = -1.03, R) >droSec1.super_0 6584930 108 + 21120651 UUACGUGC----CUACUGCAUAUUCUGUUAUUGUUUGCCA---UCGCUCGAGUGAACUUUGAUUGGCAAACAUUUCGGAAG---GCAGCCUCA--UCGUGAUCUGCAUCAACAUCUCCAU (((((...----...((((...(((((....(((((((((---(((...(((....)))))).)))))))))...))))).---)))).....--.)))))................... ( -31.30, z-score = -2.48, R) >droSim1.chr3R 13549003 108 + 27517382 UUACGUGC----CUACUGCAUAUUCUGUUAUUGUUUGCCA---UCGCUCGAGUGAACUUUGAUUGGCAAACAUUUCGGAAG---GCAGCCUCA--UCGUGAUCUGCAUCAACAUCUCCAU (((((...----...((((...(((((....(((((((((---(((...(((....)))))).)))))))))...))))).---)))).....--.)))))................... ( -31.30, z-score = -2.48, R) >consensus UUACGUGC____CUACUGCAUAUUCUGUUAUUGUUUGCCA___UCGCUCGAGUGAACUUUGAUUGGCAAACAUUUCGGAAG___GCAGCCUCA__UCGUGAUCUGCAUCAACAUCUCCAU (((((..........((((...(((((....(((((((((...(((...(((....)))))).)))))))))...)))))....))))........)))))................... (-21.14 = -22.67 + 1.53)


| Location | 7,429,200 – 7,429,313 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.72 |
| Shannon entropy | 0.26673 |
| G+C content | 0.42533 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.572639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
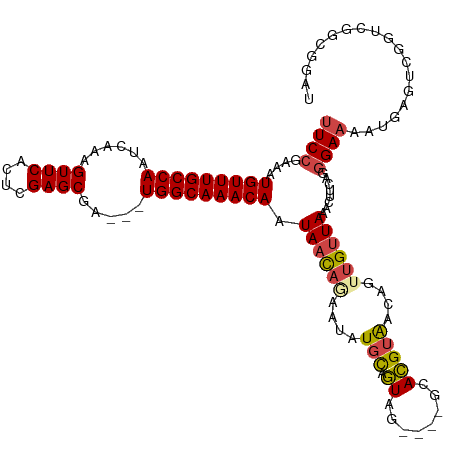
>dm3.chr3R 7429200 113 + 27905053 UUCCGAAAUGUUUGCCAAUCAAAGUUCACUCGAGCGA---UGGCAAACAAUAACAGAAUGUGCAGUAG----GCACGUAACAGUUGUUAAACUUCAGGGAAAAUGAGUCGGUCGGCGAAU (((((((.(((((((((.((...((((....))))))---))))))))).((((((.((((((.....----)))))).....))))))...))).))))......(((....))).... ( -31.60, z-score = -2.15, R) >droAna3.scaffold_13340 13874045 99 + 23697760 UUCGGAAAUGUUUGCCAAUCAAAGUUCACUCGAGCGACGGUGGCAAACAAUAAUU-AAUGUCUAAUGGAAUGGCAUGUGACAUUUGAUAAACUUGAGAGA-------------------- (((((...(((((((((......((((....)))).....)))))))))...(((-((((((..(((......)))..)))).)))))....)))))...-------------------- ( -24.50, z-score = -1.63, R) >droEre2.scaffold_4770 3555390 113 - 17746568 UUCCGAAAUGUUUGCCAAUCAAAGUUCACUCGAGCGG---UGGCAAACAAUAACAAAAUAUGCAGUAG----GCACGUAACAGAUGUUAAACUUCAGGGAAAAUGACUCGGUCGGCGGAU .(((....(((((((((......((((....))))..---)))))))))............((....(----((.((((((....))))....(((.......)))..))))).))))). ( -26.60, z-score = -0.89, R) >droYak2.chr3R 11838197 113 - 28832112 UUCCGAAAUGUUUGCCAAUCAAAGUUCACUCGAGCGG---UGGCAAACAAUAACAAAAUAUGCAGUAG----GCACGUAACAGUUGUUAAACUUCAGGGAAAAUGAGUCGGUCGGCGGAU .((((((.(((((((((......((((....))))..---))))))))).((((((.(..(((.....----)))..).....))))))...)))...........(((....)))))). ( -27.90, z-score = -1.24, R) >droSec1.super_0 6584965 113 - 21120651 UUCCGAAAUGUUUGCCAAUCAAAGUUCACUCGAGCGA---UGGCAAACAAUAACAGAAUAUGCAGUAG----GCACGUAACUGUUGUUAAACUUCAGGGAAAAUGAGUCGGUCGGCGGAU .((((((..((((((((.((...((((....))))))---))))))))((((((((.(..(((.....----)))..)..))))))))....)))...........(((....)))))). ( -30.40, z-score = -1.69, R) >droSim1.chr3R 13549038 113 - 27517382 UUCCGAAAUGUUUGCCAAUCAAAGUUCACUCGAGCGA---UGGCAAACAAUAACAGAAUAUGCAGUAG----GCACGUAACAGUUGUUAAACUUCAGGGAAAAUGACUCGGUCGGCGGAU .(((....(((((((((.((...((((....))))))---)))))))))............((....(----((.((....(((..((...(.....)...))..)))))))).))))). ( -27.00, z-score = -0.86, R) >consensus UUCCGAAAUGUUUGCCAAUCAAAGUUCACUCGAGCGA___UGGCAAACAAUAACAGAAUAUGCAGUAG____GCACGUAACAGUUGUUAAACUUCAGGGAAAAUGAGUCGGUCGGCGGAU ((((....(((((((((......((((....)))).....))))))))).((((((....(((.((........)))))....))))))........))))................... (-21.83 = -22.22 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:05 2011