
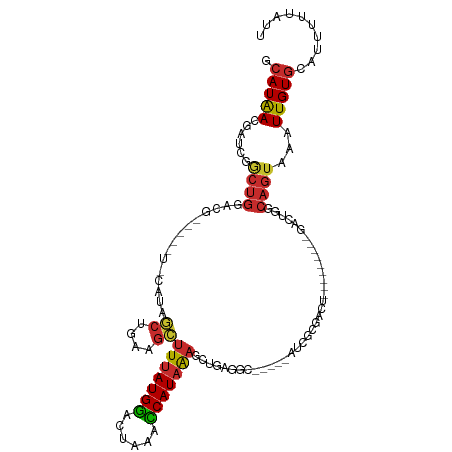
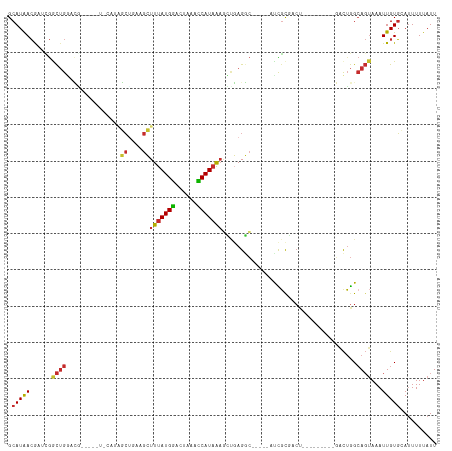
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,424,728 – 7,424,852 |
| Length | 124 |
| Max. P | 0.634529 |

| Location | 7,424,728 – 7,424,832 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.43 |
| Shannon entropy | 0.58621 |
| G+C content | 0.43508 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -8.01 |
| Energy contribution | -7.92 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.569937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7424728 104 - 27905053 GCAUAACGAUCGGCUGGACGGACUAUACAUAGCUGAAGCUUUAUGGACUAAACCAUAAAGCUGAGGC-----AUCGCGACU---------GACUGGCAGUAAAUUGUGCAUUUUUAUU ((((((......((..(.(((..........(((..((((((((((......))))))))))..)))-----.......))---------).)..))......))))))......... ( -31.63, z-score = -3.52, R) >droWil1.scaffold_181089 7803581 89 - 12369635 GCAUGAAGUGAAAUGGAAUU-------UGUUAAUGCUGCUUUAUGAACU-AAUCAUUGUCAAGAGAG-----GUUGC-------------CAUUAGAA--AAAUUGUUGA-UUUUAUU ......((((((((.((..(-------(.(((((((.(((((.(((.(.-.......))))...)))-----)).).-------------))))))..--.))...)).)-))))))) ( -12.00, z-score = 1.15, R) >droPer1.super_6 4812690 101 + 6141320 GCAUGACA--UGGCUGCUGC---CUUGCCUGCCUCAUGGUUUAUGGACC-AAUCAUAAAAGCUAGUC-----UCCGCUCCCC------UCCGCUGGCAGUAAAUUGUGGAUUUUUAUU (((.(.((--.(((....))---).))).))).(((..((((((.((..-..))......((((((.-----..........------...)))))).))))))..)))......... ( -22.04, z-score = 0.97, R) >dp4.chr2 4796925 101 + 30794189 GCAUGACA--UGGCUGCUGC---CUUGCCUGCCUCAUGGUUUAUGGACC-AAUCAUAAAAGCUAGUC-----UCCGCUCCCC------UCCGCUGGCAGUAAAUUGUGGAUUUUUAUU (((.(.((--.(((....))---).))).))).(((..((((((.((..-..))......((((((.-----..........------...)))))).))))))..)))......... ( -22.04, z-score = 0.97, R) >droAna3.scaffold_13340 13870085 109 - 23697760 GCAUAACGAUGGACUGGACC---------UGGCGAAUGCUUUAUGGACUAAACCAUAAAGUUGAGGCCUCCGACUCUGACUCUCUGGCUCGACGGGCAGUAAAUUGUGCAUUUUUAUU ((((((....(((.((((..---------.(((....(((((((((......)))))))))....)))))))..))).(((.((((......)))).)))...))))))......... ( -30.30, z-score = -1.86, R) >droEre2.scaffold_4770 3551028 94 + 17746568 GCAUAACGAUCGGCUGGACA----------AGCUGAAGCUUUAUGGACUAAACCAUAAAGCUGAGGC-----AUCGCGACU---------GACUGGCAGUAAAUUGUGCAUUUUUAUU (((((((((((((((.....----------))))))((((((((((......)))))))))).....-----.)))..(((---------(.....))))...))))))......... ( -31.60, z-score = -3.85, R) >droYak2.chr3R 11833602 98 + 28832112 GCAUAACGAUCGACUGGACGCA------GUCGCCGAAGCUUUAUGGACUAAACCAUAAAGCUGAGGC-----AUCGCGACU---------GACUGGCAGUAAAUUGUGCAUUUUUAUU ((((((((((.(((((....))------)))(((..((((((((((......))))))))))..)))-----))))..(((---------(.....))))...))))))......... ( -39.20, z-score = -5.54, R) >droSec1.super_0 6580542 104 + 21120651 GCAUAACGAUCGGCUGGACGGACUAUACAUAGCUGAAGCUUUAUGGACUAAACCAUAAAGCUGAGGC-----AUCGCGACU---------GACUGGCAGUAAAUUGUGAAUUUUUAUU .....(((((..((((..(((..........(((..((((((((((......))))))))))..)))-----.(((....)---------))))).))))..)))))........... ( -29.90, z-score = -3.39, R) >droSim1.chr3R 13544618 104 + 27517382 GCAUAACGAUCGGCUGGACGGACUAUACAUAGCUAAAGCUUUAUGGACUAAACCAUAAAGCUGAGGC-----AUCGCGACU---------GACUGGCAGUAAAUUGUGCAUUUUUAUU ((((((......((..(.(((..........(((..((((((((((......))))))))))..)))-----.......))---------).)..))......))))))......... ( -30.63, z-score = -3.35, R) >consensus GCAUAACGAUCGGCUGGACG_____U_CAUAGCUGAAGCUUUAUGGACUAAACCAUAAAGCUGAGGC_____AUCGCGACU_________GACUGGCAGUAAAUUGUGCAUUUUUAUU .(((((......((((...............((....))(((((((......))))))).....................................))))...))))).......... ( -8.01 = -7.92 + -0.08)

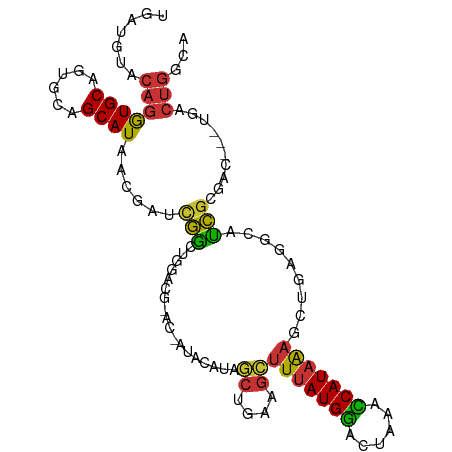
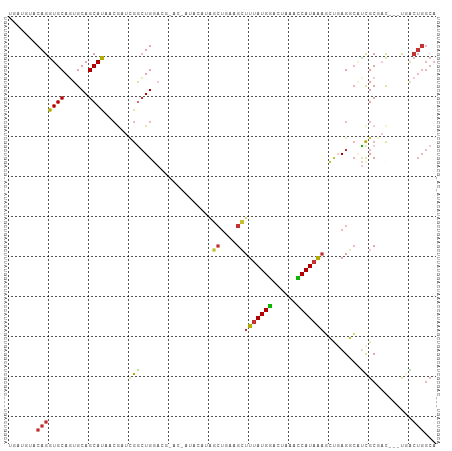
| Location | 7,424,748 – 7,424,852 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 68.69 |
| Shannon entropy | 0.62805 |
| G+C content | 0.50037 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -10.16 |
| Energy contribution | -9.54 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7424748 104 - 27905053 UGAUGUACAGGUGCAGUGCAGCAUAACGAUCGGCUGGACGGACUAUACAUAGCUGAAGCUUUAUGGACUAAACCAUAAAGCUGAGGCAUCGCGAC---UGACUGGCA ....((.(((.(((.((.((((..........)))).))............(((..((((((((((......))))))))))..)))...))).)---))))..... ( -34.50, z-score = -2.72, R) >droWil1.scaffold_181089 7803600 90 - 12369635 GAGUCCGCAGAUGCAUUGCAGCAUGAAG-UGAAAUGGA------AUUUGUUAAUGCUGCUUUAUGAACU-AAUCAUUGUCAAGAGAGGUUGCCAU---UAG------ ......((.....(((.(((((((..((-..((.....------.))..)).)))))))...)))((((-..((........))..))))))...---...------ ( -17.30, z-score = 0.97, R) >droPer1.super_6 4812710 101 + 6141320 CGAUGUACAGGUGCAGUGCAGCAUGACA--UGGCUGCU---GCCUUGCCUGCCUCAUGGUUUAUGGACC-AAUCAUAAAAGCUAGUCUCCGCUCCCCUCCGCUGGCA .(((...((((((.((.((((((.(...--...)))))---)))))))))).....(((((....))))-))))......((((((..............)))))). ( -29.14, z-score = 0.71, R) >dp4.chr2 4796945 101 + 30794189 CGAUGUACAGGUGCAGUGCAGCAUGACA--UGGCUGCU---GCCUUGCCUGCCUCAUGGUUUAUGGACC-AAUCAUAAAAGCUAGUCUCCGCUCCCCUCCGCUGGCA .(((...((((((.((.((((((.(...--...)))))---)))))))))).....(((((....))))-))))......((((((..............)))))). ( -29.14, z-score = 0.71, R) >droEre2.scaffold_4770 3551048 89 + 17746568 -----UACAGGUGCAGUGCAGCAUAACGAUCGGCUGGACA----------AGCUGAAGCUUUAUGGACUAAACCAUAAAGCUGAGGCAUCGCGAC---UGACUGGCA -----..(((.(((.((((..(.......((((((.....----------))))))((((((((((......)))))))))))..)))).))).)---))....... ( -34.30, z-score = -3.87, R) >droYak2.chr3R 11833622 93 + 28832112 -----UACAGGUGCAGUGCAGCAUAACGAUCGACUGGACGCA------GUCGCCGAAGCUUUAUGGACUAAACCAUAAAGCUGAGGCAUCGCGAC---UGACUGGCA -----..(((...((((((............(((((....))------)))(((..((((((((((......))))))))))..)))...)).))---)).)))... ( -39.20, z-score = -4.54, R) >droSec1.super_0 6580562 104 + 21120651 UGAUGUACAGGUGCACUGCAGCAUAACGAUCGGCUGGACGGACUAUACAUAGCUGAAGCUUUAUGGACUAAACCAUAAAGCUGAGGCAUCGCGAC---UGACUGGCA ....((.(((.(((..(((..(.......(((((((...(.......).)))))))((((((((((......)))))))))))..)))..))).)---))))..... ( -31.70, z-score = -2.29, R) >droSim1.chr3R 13544638 104 + 27517382 UGAUGUACAGGUGCAGUGCAGCAUAACGAUCGGCUGGACGGACUAUACAUAGCUAAAGCUUUAUGGACUAAACCAUAAAGCUGAGGCAUCGCGAC---UGACUGGCA ....((.(((.(((.((.((((..........)))).))............(((..((((((((((......))))))))))..)))...))).)---))))..... ( -33.50, z-score = -2.61, R) >consensus UGAUGUACAGGUGCAGUGCAGCAUAACGAUCGGCUGGACG_AC_AUACAUAGCUGAAGCUUUAUGGACUAAACCAUAAAGCUGAGGCAUCGCGAC___UGACUGGCA .......(((((((......))))......(((..................((....))(((((((......))))))).........)))..........)))... (-10.16 = -9.54 + -0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:03 2011