
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,416,136 – 7,416,276 |
| Length | 140 |
| Max. P | 0.956484 |

| Location | 7,416,136 – 7,416,239 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.11 |
| Shannon entropy | 0.14010 |
| G+C content | 0.30076 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.43 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
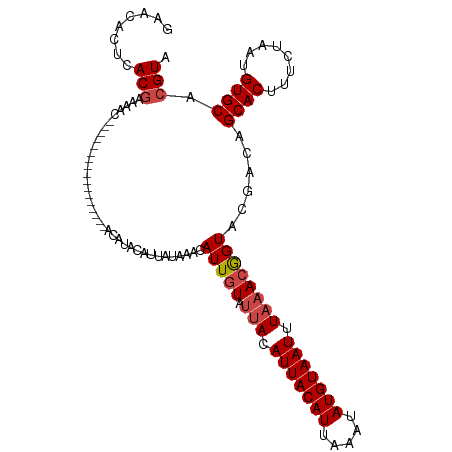
>dm3.chr3R 7416136 103 - 27905053 -------------ACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACGGUACGACAGCACUUUCUAAUGUGCACGUGCUAAUUAAGUGUAAAAAUACAAAAUUCAAGCGCGAA -------------..(((((.(((.(((((((.....))))))).))))))))......((((........)))).((((((.......((((....)))).......)))))).. ( -24.04, z-score = -2.54, R) >droAna3.scaffold_13340 13861665 103 - 23697760 -------------ACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACAGUACGCCUGCACGUACUAAUGUGCACGUACUAAUUAAGUGUAAAAAUACAAAAUUCAAGCGCGAA -------------...(((((((....(((((.....)))))((((((..((((((..(((((((....)))))))))))))..)))))).....))))))).............. ( -24.60, z-score = -2.64, R) >droEre2.scaffold_4770 3542573 102 + 17746568 -------------ACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACGGUACGCCCGCACUUUCUAAUGUGCACGUACUAAUUAAGUGU-AAAAUACAAAAUUCAAGCGCGAA -------------...(((((((....(((((.....)))))((((((..((((((...((((........)))).))))))..))))))..-..))))))).............. ( -21.80, z-score = -1.99, R) >droYak2.chr3R 11824819 103 + 28832112 -------------ACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACGGUACGCCAGCACUUUCUAAUGUGCACGUACUAAUUAAGUGUAAAAAUACAAAAUUCAAGCGCGAA -------------...(((((((....(((((.....)))))((((((..((((((...((((........)))).))))))..)))))).....))))))).............. ( -21.80, z-score = -2.07, R) >droSec1.super_0 6572133 103 + 21120651 -------------ACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACGGUACGACAGCACUUUCUAAUGUGCACGUGCUAAUUAAGUGUAAAAAUACAAAAUUCAAGCGCGAA -------------..(((((.(((.(((((((.....))))))).))))))))......((((........)))).((((((.......((((....)))).......)))))).. ( -24.04, z-score = -2.54, R) >droSim1.chr3R_random 487469 115 - 1307089 ACAUACAUUACAUACAUUAUA-AACAUUACAUUAAAUAUGUAAUUUAAACGGUACGACAGCACUUUCUAAUGUGCACGUGCUAAUUAAGUGUAAAAAUACAAAAUUCAAGCGCGAA ..........(.(((....((-(..(((((((.....))))))))))....))).)...((((........)))).((((((.......((((....)))).......)))))).. ( -20.94, z-score = -1.56, R) >consensus _____________ACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACGGUACGACAGCACUUUCUAAUGUGCACGUACUAAUUAAGUGUAAAAAUACAAAAUUCAAGCGCGAA ................(((((((....(((((.....)))))((((((..((((((...((((........)))).))))))..)))))).....))))))).............. (-20.32 = -20.43 + 0.11)



| Location | 7,416,172 – 7,416,276 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Shannon entropy | 0.23711 |
| G+C content | 0.30831 |
| Mean single sequence MFE | -16.30 |
| Consensus MFE | -13.17 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7416172 104 - 27905053 GAACACUCACGAAAACACAUACAU---------ACAUACAUUAUAAACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACGGUACGACAGCACUUUCUAAUGUGCACGUG .......((((.............---------...............(((((.(((.(((((((.....))))))).))))))))......((((........)))).)))) ( -17.30, z-score = -2.10, R) >droAna3.scaffold_13340 13861701 96 - 23697760 GAACACACACAUAUAU-----------------ACAUACAUAAUAUACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACAGUACGCCUGCACGUACUAAUGUGCACGUA ................-----------------...............(((((.(((.(((((((.....))))))).))))))))..((.(((((((....))))))).)). ( -17.70, z-score = -1.14, R) >droEre2.scaffold_4770 3542608 96 + 17746568 GAACACUCACGAAAAC-----------------ACAUACAUUAUAAACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACGGUACGCCCGCACUUUCUAAUGUGCACGUA ........(((.....-----------------...............(((((.(((.(((((((.....))))))).))))))))......((((........)))).))). ( -15.70, z-score = -1.62, R) >droYak2.chr3R 11824855 96 + 28832112 GAACACUCACGAGAAC-----------------ACAUACAUUAUAAACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACGGUACGCCAGCACUUUCUAAUGUGCACGUA ........(((.....-----------------...............(((((.(((.(((((((.....))))))).))))))))......((((........)))).))). ( -15.60, z-score = -1.42, R) >droSec1.super_0 6572169 100 + 21120651 GAACACUCACGAAAAC----ACAU---------ACAUACAUUAUAAACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACGGUACGACAGCACUUUCUAAUGUGCACGUG .......((((.....----....---------...............(((((.(((.(((((((.....))))))).))))))))......((((........)))).)))) ( -17.30, z-score = -2.23, R) >droSim1.chr3R_random 487505 112 - 1307089 GAACACUCACGAAAACACAUACAUACAUUAUAAACAUACAUUACAUACAUUAUA-AACAUUACAUUAAAUAUGUAAUUUAAACGGUACGACAGCACUUUCUAAUGUGCACGUG .......((((................................(.(((....((-(..(((((((.....))))))))))....))).)...((((........)))).)))) ( -14.20, z-score = -1.72, R) >consensus GAACACUCACGAAAAC_________________ACAUACAUUAUAAACAUUGUAUUACAUUACAUUAAAUAUGUAAUUUAAACGGUACGACAGCACUUUCUAAUGUGCACGUA ........(((.....................................(((((.(((.(((((((.....))))))).))))))))......((((........)))).))). (-13.17 = -13.70 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:00 2011