
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,395,978 – 7,396,035 |
| Length | 57 |
| Max. P | 0.994804 |

| Location | 7,395,978 – 7,396,035 |
|---|---|
| Length | 57 |
| Sequences | 4 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 63.31 |
| Shannon entropy | 0.58306 |
| G+C content | 0.47443 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -8.25 |
| Energy contribution | -9.50 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
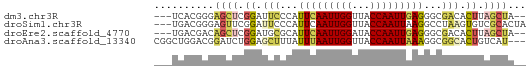
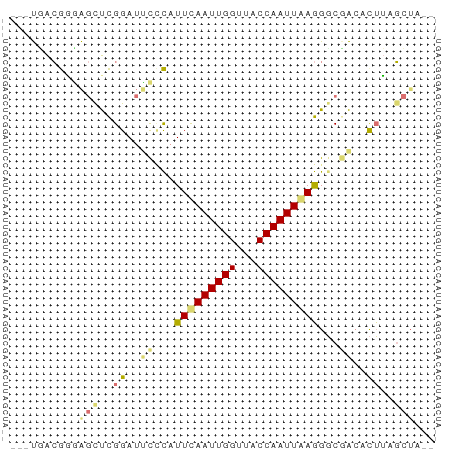
>dm3.chr3R 7395978 57 + 27905053 ---UCACGGGAGCUCGGAUUCCCAUUCAAUUGGUUACCAAUUGAGGGCGACACUUAGCUA-- ---.......((((.((..((((.(((((((((...))))))))))).))..)).)))).-- ( -17.40, z-score = -2.29, R) >droSim1.chr3R 13517352 59 - 27517382 ---UGACGGGAGUUCGGAUUCCCAUUCAAUUGGUUACCAAUUAAGGCCUAAGUGUCGCACUA ---.(((((((((....))))))....((((((...))))))...........)))...... ( -14.40, z-score = -0.75, R) >droEre2.scaffold_4770 3522164 57 - 17746568 ---UGACGACAGCUCGGAUGCGCAUUCAAUUGGAUACCAAUUGAGGGCGACACUUAGCUA-- ---.......((((.((.(((((.(((((((((...))))))))).))).)))).)))).-- ( -21.00, z-score = -4.62, R) >droAna3.scaffold_13340 13838966 59 + 23697760 CGGCUGGACGGAUCUGGAGCUUUAUUUAAUUGGUUACCAAUUAAAGGCGGCACUGUCAU--- ......(((((...((..(((...(((((((((...))))))))))))..)))))))..--- ( -15.20, z-score = -1.66, R) >consensus ___UGACGGGAGCUCGGAUUCCCAUUCAAUUGGUUACCAAUUAAGGGCGACACUUAGCUA__ ...........(((.((.(((...(((((((((...)))))))))...))).)).))).... ( -8.25 = -9.50 + 1.25)

| Location | 7,395,978 – 7,396,035 |
|---|---|
| Length | 57 |
| Sequences | 4 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 63.31 |
| Shannon entropy | 0.58306 |
| G+C content | 0.47443 |
| Mean single sequence MFE | -14.53 |
| Consensus MFE | -7.04 |
| Energy contribution | -7.72 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
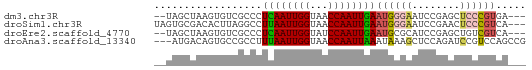
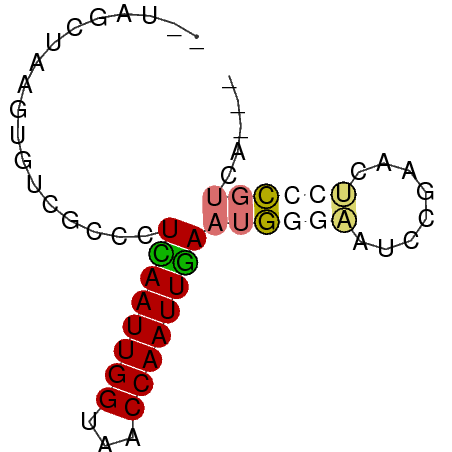
>dm3.chr3R 7395978 57 - 27905053 --UAGCUAAGUGUCGCCCUCAAUUGGUAACCAAUUGAAUGGGAAUCCGAGCUCCCGUGA--- --..........((((..((((((((...))))))))..((((........))))))))--- ( -15.60, z-score = -1.86, R) >droSim1.chr3R 13517352 59 + 27517382 UAGUGCGACACUUAGGCCUUAAUUGGUAACCAAUUGAAUGGGAAUCCGAACUCCCGUCA--- ......(((.........((((((((...))))))))..((((........))))))).--- ( -14.90, z-score = -1.30, R) >droEre2.scaffold_4770 3522164 57 + 17746568 --UAGCUAAGUGUCGCCCUCAAUUGGUAUCCAAUUGAAUGCGCAUCCGAGCUGUCGUCA--- --(((((..(((.(((..((((((((...))))))))..))))))...)))))......--- ( -20.20, z-score = -4.29, R) >droAna3.scaffold_13340 13838966 59 - 23697760 ---AUGACAGUGCCGCCUUUAAUUGGUAACCAAUUAAAUAAAGCUCCAGAUCCGUCCAGCCG ---......(.((....(((((((((...)))))))))....)).)................ ( -7.40, z-score = -0.30, R) >consensus __UAGCUAAGUGUCGCCCUCAAUUGGUAACCAAUUGAAUGGGAAUCCGAACUCCCGUCA___ ..................((((((((...))))))))((((((........))))))..... ( -7.04 = -7.72 + 0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:58 2011