
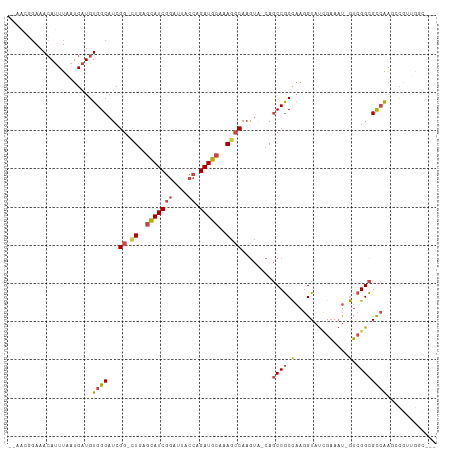
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,392,918 – 7,393,022 |
| Length | 104 |
| Max. P | 0.738977 |

| Location | 7,392,918 – 7,393,022 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.64 |
| Shannon entropy | 0.38868 |
| G+C content | 0.48978 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -17.11 |
| Energy contribution | -17.37 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7392918 104 + 27905053 --AAAUUAAAAAUUUAAUGAUGUGGCAUCGG-CUGAGCAUCGGAUUACCAGAUGCAAAGCCAAGUA-CAGCCGCCAAGGUAUCGAAAU-GUCGGCGCCAAGCCGUUGGC--- --...........((((((...((((...((-((..(((((((....)).)))))..)))).....-..((((.((...........)-).))))))))...)))))).--- ( -32.60, z-score = -1.76, R) >droAna3.scaffold_13340 13836101 108 + 23697760 AAAGCGGAAACAUUUAAUGAUGUGGCAUCGAUCUGAAGAUCAUCUUACCAGAUCCAGAUGCAAA----ACCCACUCAAGUACAGUACCCAAGGGUGUAGGAUCGCUGACGCC ..(((((..(((((....))))).((((((((((((((.....)))..))))))..)))))...----.............(..((((....))))..)..)))))...... ( -27.10, z-score = -1.16, R) >droEre2.scaffold_4770 3519013 97 - 17746568 --AGCGGAAACAUUUAAUGAUGUGGCAUAGG-UUGAGCAUCGGAUUACCAGAUGCAAAGCCAAGUUUCAGCCGCCAAGGUAUCGAAAU-GUCGGCGCCAAG----------- --.((((..(((((....)))))......((-((..(((((((....)).)))))..)))).........))))...(((..(((...-.)))..)))...----------- ( -30.70, z-score = -2.32, R) >droYak2.chr3R 11801219 96 - 28832112 --AGCGGAAACAUUUAAUGAUGUGGCAUCGG-CUGAGCAUCGGAUUACCAGAUGCAAAGCCAAGUA-CAGCCGCCAAGGUAUCGAAAU-GUCGGCGCCAAG----------- --.((((..(((((....)))))......((-((..(((((((....)).)))))..)))).....-...))))...(((..(((...-.)))..)))...----------- ( -33.20, z-score = -2.58, R) >droSec1.super_0 6548462 104 - 21120651 --AACUGAAACAUUUAAUGAUGUGGCAUCGG-CUGAGCAUCGGAUUACCAGAUGCAAAGCCAAGUA-CAGCCGCCAAGGUAUUGAAAU-GUCGGCGCCAAUCCGUUGGC--- --..((((....(((((((...((((...((-((..(((((((....)).)))))..))))..(..-...).))))...)))))))..-.)))).(((((....)))))--- ( -33.80, z-score = -1.88, R) >droSim1.chr3R 13514489 104 - 27517382 --AACUGAAACACUUAAUGAUGUGGCAUCGG-CUGAGCAUCGGAUUACCAGAUGCAAAGCCAAGUA-CAGCCGCCAAGGUAUCGAAAU-GUCGGCGCCAAUCCGUUGGC--- --...........((((((...((((...((-((..(((((((....)).)))))..)))).....-..((((.((...........)-).))))))))...)))))).--- ( -32.50, z-score = -1.37, R) >consensus __AACGGAAACAUUUAAUGAUGUGGCAUCGG_CUGAGCAUCGGAUUACCAGAUGCAAAGCCAAGUA_CAGCCGCCAAGGUAUCGAAAU_GUCGGCGCCAAGCCGUUGGC___ ......................((((...((.((..(((((((....)).)))))..))))........(((((....))...((.....)))))))))............. (-17.11 = -17.37 + 0.26)


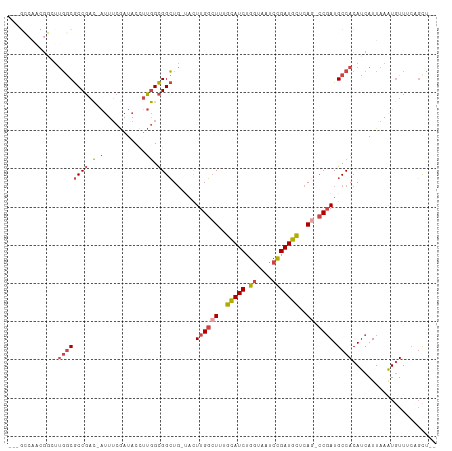
| Location | 7,392,918 – 7,393,022 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.64 |
| Shannon entropy | 0.38868 |
| G+C content | 0.48978 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -16.73 |
| Energy contribution | -17.29 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.738977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7392918 104 - 27905053 ---GCCAACGGCUUGGCGCCGAC-AUUUCGAUACCUUGGCGGCUG-UACUUGGCUUUGCAUCUGGUAAUCCGAUGCUCAG-CCGAUGCCACAUCAUUAAAUUUUUAAUUU-- ---(((((....)))))(((((.-...........)))))(((..-...((((((..(((((.((....)))))))..))-)))).))).....................-- ( -32.70, z-score = -2.37, R) >droAna3.scaffold_13340 13836101 108 - 23697760 GGCGUCAGCGAUCCUACACCCUUGGGUACUGUACUUGAGUGGGU----UUUGCAUCUGGAUCUGGUAAGAUGAUCUUCAGAUCGAUGCCACAUCAUUAAAUGUUUCCGCUUU ((((((.((((....((.((((..((((...))))..)).))))----.)))).....(((((((..((.....)))))))))))))))((((......))))......... ( -32.80, z-score = -1.66, R) >droEre2.scaffold_4770 3519013 97 + 17746568 -----------CUUGGCGCCGAC-AUUUCGAUACCUUGGCGGCUGAAACUUGGCUUUGCAUCUGGUAAUCCGAUGCUCAA-CCUAUGCCACAUCAUUAAAUGUUUCCGCU-- -----------...((((..(((-((((.(((....(((((((..(...)..)))..(((((.((....)))))))....-.....))))....))))))))))..))))-- ( -26.70, z-score = -1.90, R) >droYak2.chr3R 11801219 96 + 28832112 -----------CUUGGCGCCGAC-AUUUCGAUACCUUGGCGGCUG-UACUUGGCUUUGCAUCUGGUAAUCCGAUGCUCAG-CCGAUGCCACAUCAUUAAAUGUUUCCGCU-- -----------...((((..(((-((((.(((....(((((....-...((((((..(((((.((....)))))))..))-)))))))))....))))))))))..))))-- ( -31.81, z-score = -2.42, R) >droSec1.super_0 6548462 104 + 21120651 ---GCCAACGGAUUGGCGCCGAC-AUUUCAAUACCUUGGCGGCUG-UACUUGGCUUUGCAUCUGGUAAUCCGAUGCUCAG-CCGAUGCCACAUCAUUAAAUGUUUCAGUU-- ---(((((.(((((((.......-...))))).)))))))(((..-...((((((..(((((.((....)))))))..))-)))).)))((((......)))).......-- ( -32.80, z-score = -1.82, R) >droSim1.chr3R 13514489 104 + 27517382 ---GCCAACGGAUUGGCGCCGAC-AUUUCGAUACCUUGGCGGCUG-UACUUGGCUUUGCAUCUGGUAAUCCGAUGCUCAG-CCGAUGCCACAUCAUUAAGUGUUUCAGUU-- ---(((((....)))))(((((.-...........)))))(((((-(((((((((..(((((.((....)))))))..))-))((((...))))...)))))...)))))-- ( -34.10, z-score = -1.65, R) >consensus ___GCCAACGGCUUGGCGCCGAC_AUUUCGAUACCUUGGCGGCUG_UACUUGGCUUUGCAUCUGGUAAUCCGAUGCUCAG_CCGAUGCCACAUCAUUAAAUGUUUCAGCU__ .............(((((((((.............))))).........((((((..(((((.((....)))))))..)).)))).))))...................... (-16.73 = -17.29 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:56 2011