| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,377,588 – 7,377,715 |
| Length | 127 |
| Max. P | 0.920856 |

| Location | 7,377,588 – 7,377,682 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.71 |
| Shannon entropy | 0.16120 |
| G+C content | 0.37157 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -13.29 |
| Energy contribution | -14.09 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
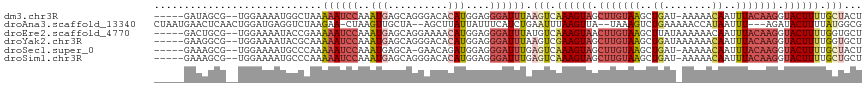
>dm3.chr3R 7377588 94 + 27905053 CUAACAUAUUUCCAGUGAGUAGCAAAAGUACCUUGUAAAUUGUUUUU-AUCAGCUUACAAGCUACUUUGACUUAAAUCCCUCCAUGUGUCCCUGC ...((((((......(((((....((((((.(((((((.(((.....-..))).))))))).)))))).))))).........))))))...... ( -19.46, z-score = -2.82, R) >droEre2.scaffold_4770 3503431 95 - 17746568 CAAACAUAUUUCCAGAAAGCACCAAAAGUACCUUGUAAAUUGUUUUUUAUAAGCUUACAAGUUACUUUGACAUAAAUCCCUCCAUGUUUUCCUGC ............(((((((((...((((((.(((((((.((((.....))))..))))))).))))))((......))......)))))).))). ( -16.80, z-score = -3.00, R) >droYak2.chr3R 11417614 95 + 28832112 CAAACAUAUUUGCAGCCAGCACCAAAAGUACCUUGUAAAUUGUUUUUUAUCAGCUUACAAGCUACUUCGACUUAAAUCCCUCCAUGUGUCCCUGC ...........((((...((((...(((((.(((((((.(((........))).))))))).))))).((......)).......))))..)))) ( -17.50, z-score = -2.41, R) >droSec1.super_0 6533191 93 - 21120651 CUUACAUAUUUCCAGUGAGUAGCAAAAGUACCUUGUAAAUUGUUUUU-AUCAGCUUACAAGCUACUUUGACUCAAAUCCCUCCAUCUGUUC-UGC ...............(((((....((((((.(((((((.(((.....-..))).))))))).)))))).))))).................-... ( -19.20, z-score = -2.86, R) >droSim1.chr3R 13498939 94 - 27517382 CUUACAUAUUUCCAGUGAGCAGCAAAAGUACCUUGUAAAUUGUUUUU-AUCAGCUUACAAGCUACUUUGACUCAAAUCCCUCCAUGUGUCCCUGC ...((((((......((((...((((.(((.(((((((.(((.....-..))).))))))).))))))).)))).........))))))...... ( -18.56, z-score = -2.23, R) >consensus CUAACAUAUUUCCAGUGAGCAGCAAAAGUACCUUGUAAAUUGUUUUU_AUCAGCUUACAAGCUACUUUGACUUAAAUCCCUCCAUGUGUCCCUGC ...((((((...............((((((.(((((((.(((........))).))))))).))))))...............))))))...... (-13.29 = -14.09 + 0.80)


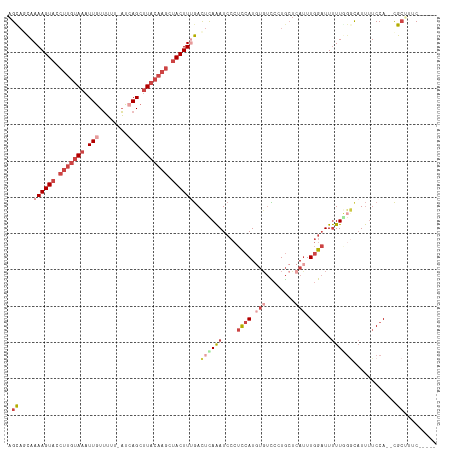
| Location | 7,377,605 – 7,377,715 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.47 |
| Shannon entropy | 0.43637 |
| G+C content | 0.36610 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -10.15 |
| Energy contribution | -13.40 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7377605 110 + 27905053 AGUAGCAAAAGUACCUUGUAAAUUGUUUUU-AUCAGCUUACAAGCUACUUUGACUUAAAUCCCUCCAUGUGUCCCUGCUCAUUUGGAUUUUUAGCCAUUUUCCA--CGCUAUC----- .(((((.((((((.(((((((.(((.....-..))).))))))).))))))(.((.((((((....(((.((....)).)))..))))))..)).)........--.))))).----- ( -24.40, z-score = -3.60, R) >droAna3.scaffold_13340 13821986 110 + 23697760 CGCCAUAAAAGUAUCU---AAAUUAUGGUUUUUCAGACUUUA--UAACUUAAAUUCAGAUGAAAUAAUAAGCU--UAGCAACUUAG-UUCUUAGACCUCAUCCAGUUGAGUUCAUUAG .(((((((........---...))))))).............--.(((((((.....(((((.....((((..--.(((......)-))))))....)))))...)))))))...... ( -14.70, z-score = 0.13, R) >droEre2.scaffold_4770 3503448 111 - 17746568 AGCACCAAAAGUACCUUGUAAAUUGUUUUUUAUAAGCUUACAAGUUACUUUGACAUAAAUCCCUCCAUGUUUUCCUGCUCAUUUGGAUUUUUCGGUAUUUUCCA--CGCAGUC----- .((....((((((.(((((((.((((.....))))..))))))).))))))((...((((((......((......))......)))))).))((......)).--.))....----- ( -21.60, z-score = -2.63, R) >droYak2.chr3R 11417631 111 + 28832112 AGCACCAAAAGUACCUUGUAAAUUGUUUUUUAUCAGCUUACAAGCUACUUCGACUUAAAUCCCUCCAUGUGUCCCUGCUCAUUUGGAUUUUUGCGUAUUUUCCA--CGCCUUC----- .((.....(((((.(((((((.(((........))).))))))).)))))((.(..((((((....(((.((....)).)))..))))))..))).........--.))....----- ( -21.20, z-score = -2.92, R) >droSec1.super_0 6533208 109 - 21120651 AGUAGCAAAAGUACCUUGUAAAUUGUUUUU-AUCAGCUUACAAGCUACUUUGACUCAAAUCCCUCCAUCUGUUC-UGCUCAUUUGGAUUUUUGGGCAUUUUCCA--CGCUUUC----- ...(((.((((((.(((((((.(((.....-..))).))))))).))))))(.((((((....((((..((...-....))..))))..)))))))........--.)))...----- ( -24.80, z-score = -2.75, R) >droSim1.chr3R 13498956 110 - 27517382 AGCAGCAAAAGUACCUUGUAAAUUGUUUUU-AUCAGCUUACAAGCUACUUUGACUCAAAUCCCUCCAUGUGUCCCUGCUCAUUUGGAUUUUUGGGCAUUUUCCA--CGCUUUC----- ...(((.((((((.(((((((.(((.....-..))).))))))).))))))(.((((((....((((.(((........))).))))..)))))))........--.)))...----- ( -26.30, z-score = -2.62, R) >consensus AGCAGCAAAAGUACCUUGUAAAUUGUUUUU_AUCAGCUUACAAGCUACUUUGACUCAAAUCCCUCCAUGUGUCCCUGCUCAUUUGGAUUUUUGGGCAUUUUCCA__CGCUUUC_____ .((....((((((.(((((((.(((........))).))))))).))))))..((((((....((((.(((........))).))))..))))))............))......... (-10.15 = -13.40 + 3.25)

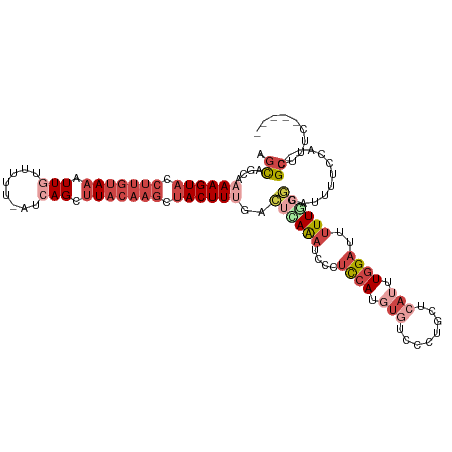
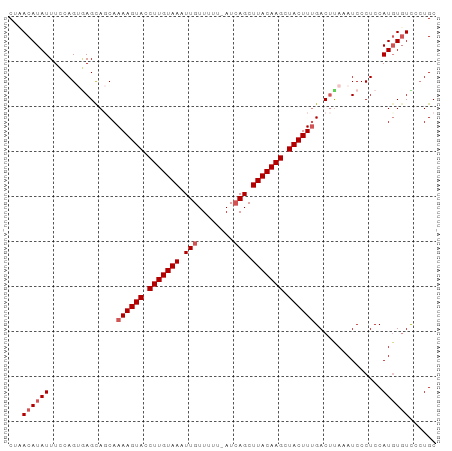
| Location | 7,377,605 – 7,377,715 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.47 |
| Shannon entropy | 0.43637 |
| G+C content | 0.36610 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -12.27 |
| Energy contribution | -14.33 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
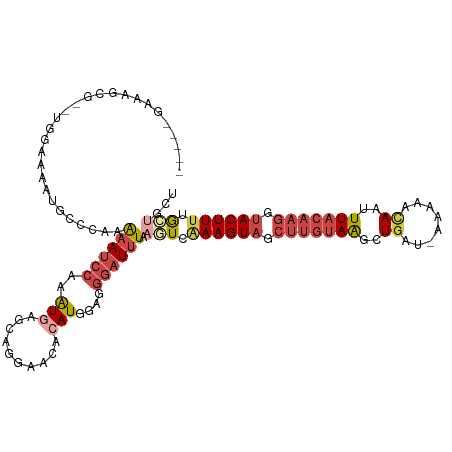
>dm3.chr3R 7377605 110 - 27905053 -----GAUAGCG--UGGAAAAUGGCUAAAAAUCCAAAUGAGCAGGGACACAUGGAGGGAUUUAAGUCAAAGUAGCUUGUAAGCUGAU-AAAAACAAUUUACAAGGUACUUUUGCUACU -----..(((((--.......(((((..((((((..(((.(......).)))....)))))).)))))(((((.((((((((.((..-.....)).)))))))).))))).))))).. ( -27.80, z-score = -2.69, R) >droAna3.scaffold_13340 13821986 110 - 23697760 CUAAUGAACUCAACUGGAUGAGGUCUAAGAA-CUAAGUUGCUA--AGCUUAUUAUUUCAUCUGAAUUUAAGUUA--UAAAGUCUGAAAAACCAUAAUUU---AGAUACUUUUAUGGCG ...............(((((((((.....((-.((((((....--)))))))))))))))))........((((--(((((((((((.........)))---))))...)))))))). ( -18.30, z-score = -0.36, R) >droEre2.scaffold_4770 3503448 111 + 17746568 -----GACUGCG--UGGAAAAUACCGAAAAAUCCAAAUGAGCAGGAAAACAUGGAGGGAUUUAUGUCAAAGUAACUUGUAAGCUUAUAAAAAACAAUUUACAAGGUACUUUUGGUGCU -----..((((.--((((.............)))).....))))....(((((((....))))))).((((((.((((((((..............)))))))).))))))....... ( -21.36, z-score = -0.92, R) >droYak2.chr3R 11417631 111 - 28832112 -----GAAGGCG--UGGAAAAUACGCAAAAAUCCAAAUGAGCAGGGACACAUGGAGGGAUUUAAGUCGAAGUAGCUUGUAAGCUGAUAAAAAACAAUUUACAAGGUACUUUUGGUGCU -----....(((--((.....)))))..((((((..(((.(......).)))....)))))).....((((((.((((((((.((........)).)))))))).))))))....... ( -26.80, z-score = -2.34, R) >droSec1.super_0 6533208 109 + 21120651 -----GAAAGCG--UGGAAAAUGCCCAAAAAUCCAAAUGAGCA-GAACAGAUGGAGGGAUUUGAGUCAAAGUAGCUUGUAAGCUGAU-AAAAACAAUUUACAAGGUACUUUUGCUACU -----....(((--(.....))))((.....((((..((....-...))..)))).)).....(((.((((((.((((((((.((..-.....)).)))))))).)))))).)))... ( -26.40, z-score = -2.14, R) >droSim1.chr3R 13498956 110 + 27517382 -----GAAAGCG--UGGAAAAUGCCCAAAAAUCCAAAUGAGCAGGGACACAUGGAGGGAUUUGAGUCAAAGUAGCUUGUAAGCUGAU-AAAAACAAUUUACAAGGUACUUUUGCUGCU -----...(((.--(((.......))).((((((..(((.(......).)))....)))))).(((.((((((.((((((((.((..-.....)).)))))))).)))))).)))))) ( -27.20, z-score = -1.36, R) >consensus _____GAAAGCG__UGGAAAAUGCCCAAAAAUCCAAAUGAGCAGGAACACAUGGAGGGAUUUAAGUCAAAGUAGCUUGUAAGCUGAU_AAAAACAAUUUACAAGGUACUUUUGCUGCU ............................((((((..(((..........)))....)))))).(((.((((((.(((((((..((........))..))))))).)))))).)))... (-12.27 = -14.33 + 2.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:52 2011