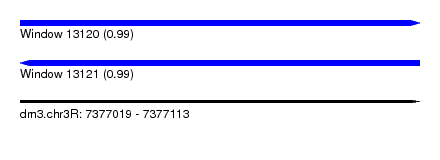
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,377,019 – 7,377,113 |
| Length | 94 |
| Max. P | 0.990841 |

| Location | 7,377,019 – 7,377,113 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.60 |
| Shannon entropy | 0.53256 |
| G+C content | 0.43192 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -14.44 |
| Energy contribution | -15.76 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.987976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
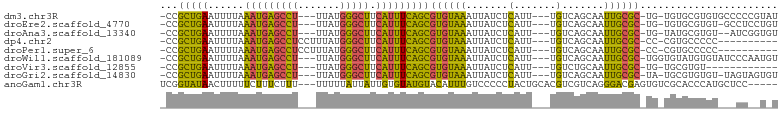
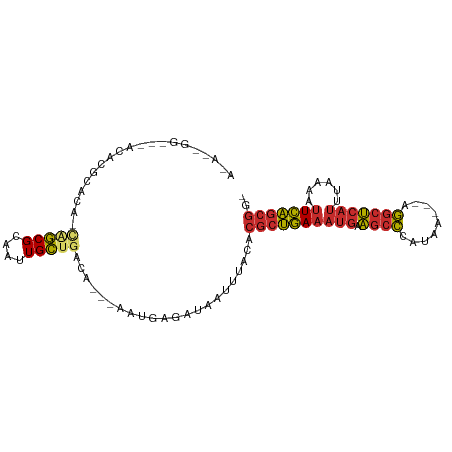
>dm3.chr3R 7377019 94 + 27905053 -CCGCUGAAUUUUAAAUGAGCCU---UUAUGGGCUUCAUUUCAGCGUGUAAAUUAUCUCAUU---UGUCAGCAAUUGCGC-UG-UGUGCGUGUGCCCCCGUAU -.(((((((......((((((((---....)))).)))))))))))................---.....(((...((((-..-...)))).)))........ ( -24.90, z-score = -1.61, R) >droEre2.scaffold_4770 3502878 93 - 17746568 -CCGCUGAAUUUUAAAUGAGCCU---UUAUGGGCUUCAUUUCAGCGUGUAAAUUAUCUCAUU---UGUCAGCAAUUGCGC-UG-UGUGCGUGU-GCCUCCUGU -.(((((((......((((((((---....)))).)))))))))))................---.....(((...((((-..-...)))).)-))....... ( -24.90, z-score = -1.65, R) >droAna3.scaffold_13340 13821282 92 + 23697760 -CCGCUGAAUUUUAAAUGAGCCU---UUAUGGGCUUCAUUUCAGCGUGUAAAUUAUCUCAUU---UGUCAGCAAUUGCGC-UG-UAUGCGUGU--AUCGGUGU -(((((((....((((((((..(---(((((.(((.......))).))))))....))))))---)))))))....((((-..-...))))..--...))... ( -25.50, z-score = -1.87, R) >dp4.chr2 25655687 87 + 30794189 -CCGCUGAAUUUUAAAUGAGCCUCCUUUAUGGGCUUCAUUUCAGCGUGUAAAUUAUCUCAUU---UGUCAGCAAUUGCGC-CC-CGUGCCCCC---------- -..(((((....((((((((.....((((((.(((.......))).))))))....))))))---)))))))....((((-..-.))))....---------- ( -21.60, z-score = -2.47, R) >droPer1.super_6 998335 87 + 6141320 -CCGCUGAAUUUUAAAUGAGCCUCCUUUAUGGGCUUCAUUUCAGCGUGUAAAUUAUCUCAUU---UGUCAGCAAUUGCGC-CC-CGUGCCCCC---------- -..(((((....((((((((.....((((((.(((.......))).))))))....))))))---)))))))....((((-..-.))))....---------- ( -21.60, z-score = -2.47, R) >droWil1.scaffold_181089 8262286 95 - 12369635 -CCGCUGAAUUUUAAAUGAGCCU---UUAUGGGCUUCAUUUCAGCGUGUAAAUUAUCUCAUU---UGUCAGCAAUUGCGC-UGGUGUAUGUGUAUCCCAAUGU -.(((((((......((((((((---....)))).)))))))))))..........(.(((.---(..((((......))-))..).))).)........... ( -25.10, z-score = -2.06, R) >droVir3.scaffold_12855 9877350 82 + 10161210 -CCGCUGAAUUUUAAAUGAGCCU---UUAUGGGCUUCAUUUCAGCGUGUAAAUUAUCUCAUU---UGUCUGCAAUUGCGC-UG-UGCGUGU------------ -.(((........((((((((((---....)))).))))))(((((((........(.....---.)........)))))-))-.)))...------------ ( -19.99, z-score = -1.06, R) >droGri2.scaffold_14830 2213741 93 + 6267026 -CCGCUGAAUUUUAAAUGAGCCU---UUAUGGGCUUCAUUUCAGCGUGUAAAUUAUCUCAUU---UGUCAGCAAUUGCGC-UA-UGCGUGUGU-UAGUAGUGU -..(((((....((((((((..(---(((((.(((.......))).))))))....))))))---)))))))....((((-((-(........-..))))))) ( -24.30, z-score = -1.50, R) >anoGam1.chr3R 25047578 95 + 53272125 UCGGUAUAACUUUUUCUUUCUUU---UUUUUAUUAUUGUGUAUGUACAUUUGUCCCCCUACUGCACGUCGUCAGGGACGAGUGUCGCACCCAUGCUCC----- .......................---...........(((..((((((((((((((...((........))..))))))))))).)))..))).....----- ( -20.30, z-score = -1.95, R) >consensus _CCGCUGAAUUUUAAAUGAGCCU___UUAUGGGCUUCAUUUCAGCGUGUAAAUUAUCUCAUU___UGUCAGCAAUUGCGC_UG_UGUGCGUGU___CC__U_U ...(((((......((((((......(((((.(((.......))).))))).....)))))).....)))))....((((.....)))).............. (-14.44 = -15.76 + 1.31)
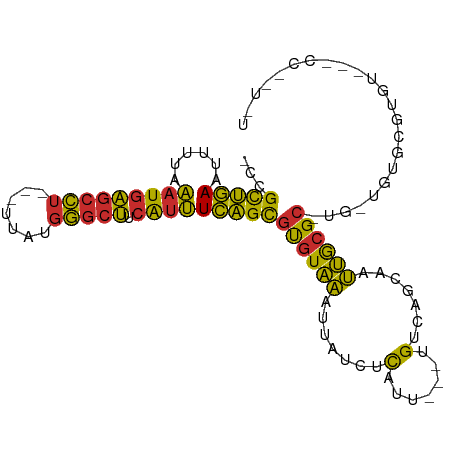
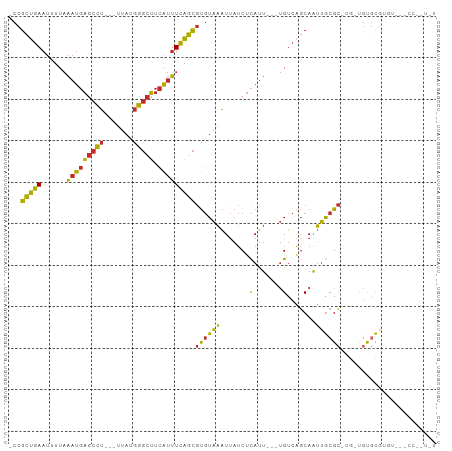
| Location | 7,377,019 – 7,377,113 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 73.60 |
| Shannon entropy | 0.53256 |
| G+C content | 0.43192 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -12.19 |
| Energy contribution | -12.21 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7377019 94 - 27905053 AUACGGGGGCACACGCACA-CA-GCGCAAUUGCUGACA---AAUGAGAUAAUUUACACGCUGAAAUGAAGCCCAUAA---AGGCUCAUUUAAAAUUCAGCGG- ....(..((((..(((...-..-)))....))))..).---................(((((((((((.(((.....---.)))))))......))))))).- ( -24.00, z-score = -2.42, R) >droEre2.scaffold_4770 3502878 93 + 17746568 ACAGGAGGC-ACACGCACA-CA-GCGCAAUUGCUGACA---AAUGAGAUAAUUUACACGCUGAAAUGAAGCCCAUAA---AGGCUCAUUUAAAAUUCAGCGG- ...(..(((-(..(((...-..-)))....))))..).---................(((((((((((.(((.....---.)))))))......))))))).- ( -24.30, z-score = -2.92, R) >droAna3.scaffold_13340 13821282 92 - 23697760 ACACCGAU--ACACGCAUA-CA-GCGCAAUUGCUGACA---AAUGAGAUAAUUUACACGCUGAAAUGAAGCCCAUAA---AGGCUCAUUUAAAAUUCAGCGG- ........--.........-((-(((....)))))...---................(((((((((((.(((.....---.)))))))......))))))).- ( -21.10, z-score = -2.82, R) >dp4.chr2 25655687 87 - 30794189 ----------GGGGGCACG-GG-GCGCAAUUGCUGACA---AAUGAGAUAAUUUACACGCUGAAAUGAAGCCCAUAAAGGAGGCUCAUUUAAAAUUCAGCGG- ----------(..((((..-(.-...)...))))..).---................(((((((((((.((((......).)))))))......))))))).- ( -19.50, z-score = -0.58, R) >droPer1.super_6 998335 87 - 6141320 ----------GGGGGCACG-GG-GCGCAAUUGCUGACA---AAUGAGAUAAUUUACACGCUGAAAUGAAGCCCAUAAAGGAGGCUCAUUUAAAAUUCAGCGG- ----------(..((((..-(.-...)...))))..).---................(((((((((((.((((......).)))))))......))))))).- ( -19.50, z-score = -0.58, R) >droWil1.scaffold_181089 8262286 95 + 12369635 ACAUUGGGAUACACAUACACCA-GCGCAAUUGCUGACA---AAUGAGAUAAUUUACACGCUGAAAUGAAGCCCAUAA---AGGCUCAUUUAAAAUUCAGCGG- .............(((....((-(((....)))))...---.)))............(((((((((((.(((.....---.)))))))......))))))).- ( -21.30, z-score = -2.45, R) >droVir3.scaffold_12855 9877350 82 - 10161210 ------------ACACGCA-CA-GCGCAAUUGCAGACA---AAUGAGAUAAUUUACACGCUGAAAUGAAGCCCAUAA---AGGCUCAUUUAAAAUUCAGCGG- ------------...(((.-((-((((....))....(---(((......))))....))))((((((.(((.....---.)))))))))........))).- ( -21.10, z-score = -3.28, R) >droGri2.scaffold_14830 2213741 93 - 6267026 ACACUACUA-ACACACGCA-UA-GCGCAAUUGCUGACA---AAUGAGAUAAUUUACACGCUGAAAUGAAGCCCAUAA---AGGCUCAUUUAAAAUUCAGCGG- .........-.........-((-(((....)))))...---................(((((((((((.(((.....---.)))))))......))))))).- ( -19.00, z-score = -2.24, R) >anoGam1.chr3R 25047578 95 - 53272125 -----GGAGCAUGGGUGCGACACUCGUCCCUGACGACGUGCAGUAGGGGGACAAAUGUACAUACACAAUAAUAAAAA---AAAGAAAGAAAAAGUUAUACCGA -----........((((..((..(((((...))))).(((((....(....)...))))).................---.............))..)))).. ( -20.20, z-score = -1.23, R) >consensus A_A__GG___ACACGCACA_CA_GCGCAAUUGCUGACA___AAUGAGAUAAUUUACACGCUGAAAUGAAGCCCAUAA___AGGCUCAUUUAAAAUUCAGCGG_ .......................(((....)))........................((((((((((.((((.........)))))))......))))))).. (-12.19 = -12.21 + 0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:50 2011