
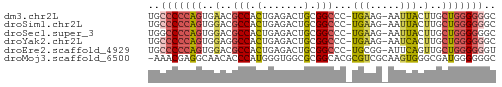
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,083,228 – 7,083,284 |
| Length | 56 |
| Max. P | 0.699265 |

| Location | 7,083,228 – 7,083,284 |
|---|---|
| Length | 56 |
| Sequences | 6 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 76.47 |
| Shannon entropy | 0.44762 |
| G+C content | 0.65863 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -12.97 |
| Energy contribution | -13.37 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
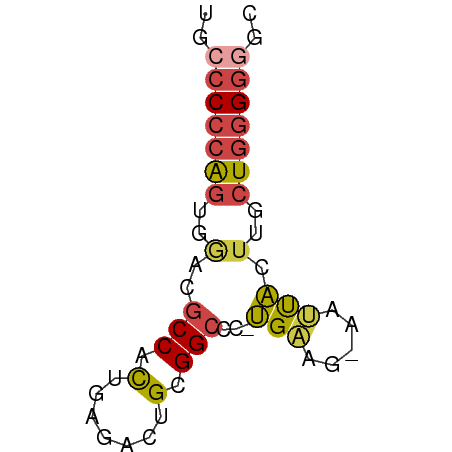
>dm3.chr2L 7083228 56 + 23011544 UGCCCCCAGUGAACGCCACUGAGACUGCGGCCC-UGAAG-AAUUACUUGCUGGGGGGC ..((((((((....(((.(.......).)))..-..(((-.....))))))))))).. ( -20.60, z-score = -1.69, R) >droSim1.chr2L 6870543 56 + 22036055 UGCCCCCAGUGGACGCCACUGAGACUGCGGCCC-UGAAG-AAUUACUUGCUGGGGGGC ..((((((((((..(((.(.......).)))))-..(((-.....))))))))))).. ( -21.20, z-score = -1.31, R) >droSec1.super_3 2607250 56 + 7220098 UGGCCCCAGUGGACGCCACUGAGACUGCGGCCC-UGAAG-AAUUACUUGCUGGGGGGC ..((((((((((...)))))).........(((-.((((-.....))).).))))))) ( -18.50, z-score = -0.13, R) >droYak2.chr2L 16498938 56 - 22324452 UGCCCCCAGUGGAGGCCACUGAGACUGCGGCCC-UGAAG-AAUCACUUGCUGGGGGGC ..(((((((..(.((((.(.......).)))).-(((..-..))).)..))))))).. ( -25.40, z-score = -1.77, R) >droEre2.scaffold_4929 15997702 56 + 26641161 UGCCCCCAGUGGACGCCACUGAGACUGCGGCCC-UGCGG-AUUCAGUUGCUGGGGGGU ..((((((((((...))((((((.(((((....-)))))-.)))))).)))))))).. ( -28.50, z-score = -2.13, R) >droMoj3.scaffold_6500 27867100 57 - 32352404 -AAACGAGGCAACACCCAUGGGUGGCGCGGCACGCGUCGCAAGUGGGCGAUGGGGGGC -........((.(.(((((..((((((((...))))))))..))))).).))...... ( -23.40, z-score = -1.70, R) >consensus UGCCCCCAGUGGACGCCACUGAGACUGCGGCCC_UGAAG_AAUUACUUGCUGGGGGGC ..(((((((..(..(((.(.......).)))...(((.....))).)..))))))).. (-12.97 = -13.37 + 0.39)
| Location | 7,083,228 – 7,083,284 |
|---|---|
| Length | 56 |
| Sequences | 6 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 76.47 |
| Shannon entropy | 0.44762 |
| G+C content | 0.65863 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -12.24 |
| Energy contribution | -13.97 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
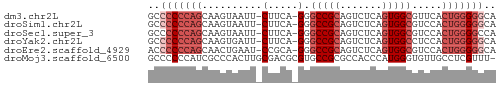
>dm3.chr2L 7083228 56 - 23011544 GCCCCCCAGCAAGUAAUU-CUUCA-GGGCCGCAGUCUCAGUGGCGUUCACUGGGGGCA ((((((..(.(((.....-)))).-..(((((.......))))).......)))))). ( -21.80, z-score = -1.53, R) >droSim1.chr2L 6870543 56 - 22036055 GCCCCCCAGCAAGUAAUU-CUUCA-GGGCCGCAGUCUCAGUGGCGUCCACUGGGGGCA ((((((..(.(((.....-)))).-(((((((.......)))))..))...)))))). ( -22.50, z-score = -1.52, R) >droSec1.super_3 2607250 56 - 7220098 GCCCCCCAGCAAGUAAUU-CUUCA-GGGCCGCAGUCUCAGUGGCGUCCACUGGGGCCA ((..(((.(.(((.....-)))).-)))..)).((((((((((...)))))))))).. ( -21.50, z-score = -1.59, R) >droYak2.chr2L 16498938 56 + 22324452 GCCCCCCAGCAAGUGAUU-CUUCA-GGGCCGCAGUCUCAGUGGCCUCCACUGGGGGCA ((((((.((....(((..-..)))-(((((((.......)))))))...)))))))). ( -27.00, z-score = -2.18, R) >droEre2.scaffold_4929 15997702 56 - 26641161 ACCCCCCAGCAACUGAAU-CCGCA-GGGCCGCAGUCUCAGUGGCGUCCACUGGGGGCA ..(((((((...(((...-...))-).(((((.......))))).....))))))).. ( -24.20, z-score = -1.14, R) >droMoj3.scaffold_6500 27867100 57 + 32352404 GCCCCCCAUCGCCCACUUGCGACGCGUGCCGCGCCACCCAUGGGUGUUGCCUCGUUU- ........((((......)))).(((.((.(((((.......))))).))..)))..- ( -15.00, z-score = 0.15, R) >consensus GCCCCCCAGCAAGUAAUU_CUUCA_GGGCCGCAGUCUCAGUGGCGUCCACUGGGGGCA ..(((((((..........((....))(((((.......))))).....))))))).. (-12.24 = -13.97 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:51 2011