
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,347,401 – 7,347,494 |
| Length | 93 |
| Max. P | 0.884062 |

| Location | 7,347,401 – 7,347,494 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 66.93 |
| Shannon entropy | 0.65262 |
| G+C content | 0.62832 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -11.71 |
| Energy contribution | -13.04 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
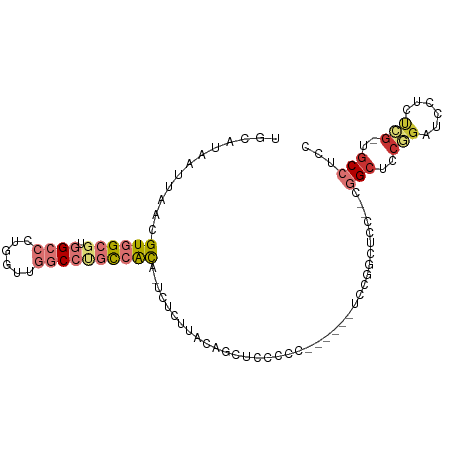
>dm3.chr3R 7347401 93 + 27905053 UGCAUAAUUAACGUGGCGUGGCCCUGGUUGGCCUGCCACA-UCUCUUACAGCUUCUCC------UCCGGCUCCUCCGGCUCCGGAUCCUCUCG-UGCCUCC .((((.......((((((.((((......)))))))))).-.................------(((((..((...))..))))).......)-))).... ( -29.80, z-score = -2.12, R) >droSim1.chr3R 13471110 84 - 27517382 UGCAUAAUUAACGUGGCGUGGCCCUGGUUGGCCUGCCACA-UCUCUUACAGC------------UCCGGCUCC---GGCUCCGGAUCCUCUCG-UGCCUCC .((((.......((((((.((((......)))))))))).-...........------------...((.(((---(....)))).))....)-))).... ( -30.80, z-score = -2.27, R) >droSec1.super_0 6503452 90 - 21120651 UGCAUAAUUAACGUGGCGUGGCCCUGGUUGGCCUGCCACA-UCUCUUACAGCUCCUCC------UCCGGCUCC---GGCUCCGGAUCCUCUCG-UGCCUCC .((((.......((((((.((((......)))))))))).-.................------...((.(((---(....)))).))....)-))).... ( -30.80, z-score = -2.37, R) >droYak2.chr3R 11382914 99 + 28832112 UGCAUAAUUAACGUGGCGUGGCCCUGGUUGGCCUGCCACA-UCUCUUACAGCUCCUCCAGCUCCUCUGGCUCCUCCGGCUCCGGAUCCGCUCG-UGCCUCC .((((.......((((((.((((......)))))))))).-........(((....((((.....))))....((((....))))...))).)-))).... ( -32.50, z-score = -1.79, R) >droEre2.scaffold_4770 3473655 81 - 17746568 UGCAUAAUUAACGUGGCGUGGCCCUGGUUGGCCUGCCACA-UCUCUUACAGCUC---------------CUCC---GGCUCCGGAUCCGCUGG-UGCCUCC .((((.......((((((.((((......)))))))))).-.......((((..---------------.(((---(....))))...)))))-))).... ( -33.90, z-score = -3.19, R) >droAna3.scaffold_13340 13791030 97 + 23697760 UGCAUAAUUAACGUGGCGUGGCCCUGGUUGGCCCGCCACA-UCUCUUACUACCGCCACUGGUGUUCCUGCCUC--CGGGUGGCAAGCCGCCCA-AGCCUCC .(((........((((((.((((......)))))))))).-........(((((....)))))....)))...--.((((((....)))))).-....... ( -37.50, z-score = -1.67, R) >dp4.Unknown_group_353 11788 100 + 17550 UGCAUAAUUAACGUGGCCCUGGCAUGUCCUGCCACUGGUGGUGGCACCCCACAGACCCCUGGCCUUCUCCUCCA-CAGCCACUGCCCCUCCAGCUGCCUCC .(((........(((((..(((........((((..(((.((((....))))..)))..))))........)))-..))))).((.......))))).... ( -28.49, z-score = 0.03, R) >droPer1.super_1368 39 100 - 7994 GUUGGGGUUAACGUGGCCCUGGCAUGUCCUGCCACUGGUGGUGGCACCCCACAGACCCCUGGCCUUCUCCGCUA-CAGCUACUGCCCCUCCAGCUGCCUCC (((((((.....(((((..((((.......((((..(((.((((....))))..)))..)))).......))))-..))))).....)))))))....... ( -35.14, z-score = 0.25, R) >consensus UGCAUAAUUAACGUGGCGUGGCCCUGGUUGGCCUGCCACA_UCUCUUACAGCUCCCCC______UCCGGCUCC__CGGCUCCGGAUCCUCUCG_UGCCUCC ............((((((.((((......)))))))))).....................................(((..(((......)))..)))... (-11.71 = -13.04 + 1.33)


| Location | 7,347,401 – 7,347,494 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 66.93 |
| Shannon entropy | 0.65262 |
| G+C content | 0.62832 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -11.87 |
| Energy contribution | -13.31 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.884062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
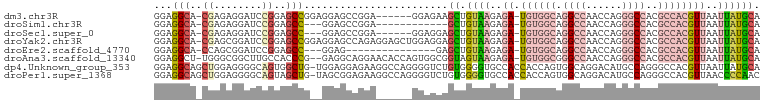
>dm3.chr3R 7347401 93 - 27905053 GGAGGCA-CGAGAGGAUCCGGAGCCGGAGGAGCCGGA------GGAGAAGCUGUAAGAGA-UGUGGCAGGCCAACCAGGGCCACGCCACGUUAAUUAUGCA ...(((.-(....)..((((....))))...)))...------......((.((((..((-((((((.((((......))))..))))))))..)))))). ( -36.20, z-score = -3.02, R) >droSim1.chr3R 13471110 84 + 27517382 GGAGGCA-CGAGAGGAUCCGGAGCC---GGAGCCGGA------------GCUGUAAGAGA-UGUGGCAGGCCAACCAGGGCCACGCCACGUUAAUUAUGCA ...(((.-(....)..((((....)---))))))...------------((.((((..((-((((((.((((......))))..))))))))..)))))). ( -36.20, z-score = -3.38, R) >droSec1.super_0 6503452 90 + 21120651 GGAGGCA-CGAGAGGAUCCGGAGCC---GGAGCCGGA------GGAGGAGCUGUAAGAGA-UGUGGCAGGCCAACCAGGGCCACGCCACGUUAAUUAUGCA ...(((.-(....)..((((....)---))))))...------......((.((((..((-((((((.((((......))))..))))))))..)))))). ( -36.20, z-score = -2.98, R) >droYak2.chr3R 11382914 99 - 28832112 GGAGGCA-CGAGCGGAUCCGGAGCCGGAGGAGCCAGAGGAGCUGGAGGAGCUGUAAGAGA-UGUGGCAGGCCAACCAGGGCCACGCCACGUUAAUUAUGCA ....((.-..(((...(((((..((...(....)...))..)))))...)))((((..((-((((((.((((......))))..))))))))..)))))). ( -39.40, z-score = -2.60, R) >droEre2.scaffold_4770 3473655 81 + 17746568 GGAGGCA-CCAGCGGAUCCGGAGCC---GGAG---------------GAGCUGUAAGAGA-UGUGGCAGGCCAACCAGGGCCACGCCACGUUAAUUAUGCA ....(((-.((((...((((....)---))).---------------..))))(((..((-((((((.((((......))))..))))))))..)))))). ( -36.20, z-score = -3.41, R) >droAna3.scaffold_13340 13791030 97 - 23697760 GGAGGCU-UGGGCGGCUUGCCACCCG--GAGGCAGGAACACCAGUGGCGGUAGUAAGAGA-UGUGGCGGGCCAACCAGGGCCACGCCACGUUAAUUAUGCA ....(((-(((....((((((.....--..))))))....)))..))).(((.(((..((-(((((((((((......)))).)))))))))..)))))). ( -42.00, z-score = -1.65, R) >dp4.Unknown_group_353 11788 100 - 17550 GGAGGCAGCUGGAGGGGCAGUGGCUG-UGGAGGAGAAGGCCAGGGGUCUGUGGGGUGCCACCACCAGUGGCAGGACAUGCCAGGGCCACGUUAAUUAUGCA ....((((((.....))).((((((.-(((........((((..(((..((((....)))).)))..))))........))).))))))........))). ( -39.69, z-score = -0.51, R) >droPer1.super_1368 39 100 + 7994 GGAGGCAGCUGGAGGGGCAGUAGCUG-UAGCGGAGAAGGCCAGGGGUCUGUGGGGUGCCACCACCAGUGGCAGGACAUGCCAGGGCCACGUUAACCCCAAC ...(((..(((..(.(((....))).-)..))).....))).(((((.(((((((((....))))..(((((.....)))))...)))))...)))))... ( -37.70, z-score = 0.74, R) >consensus GGAGGCA_CGAGAGGAUCCGGAGCCG__GGAGCCGGA______GGAGCAGCUGUAAGAGA_UGUGGCAGGCCAACCAGGGCCACGCCACGUUAAUUAUGCA ...(((..((........))..)))........................((.((((.....((((((.((((......))))..))))))....)))))). (-11.87 = -13.31 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:43 2011