| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,345,331 – 7,345,431 |
| Length | 100 |
| Max. P | 0.993682 |

| Location | 7,345,331 – 7,345,431 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.13 |
| Shannon entropy | 0.53081 |
| G+C content | 0.52871 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -18.87 |
| Energy contribution | -20.53 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
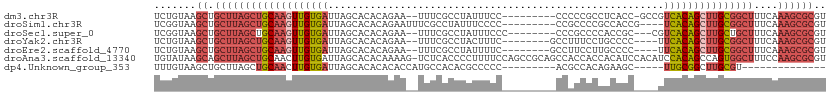
>dm3.chr3R 7345331 100 + 27905053 UCUGUAAGCUGCUUAGCUGCAAGUUGUGAUUAGCACACAGAA--UUUCGCCUAUUUCC---------CCCCCGCCUCACC-GCCGUCACAGCUUGCGGCUUUCAAAGCGCGU .......((.((((((((((((((((((((..((.....(..--...)((........---------.....))......-)).))))))))))))))))....)))))).. ( -31.02, z-score = -2.67, R) >droSim1.chr3R 13469068 99 - 27517382 UCGGUAAGCUGCUUAGCUGCAAGUUGUGAUUAGCACACAGAAUUUCGCCUAUUUCCCC---------CCGCCCCGCCACCG----UCACAGCUUGCGGCUUUCAAAGCGCGU .......((.((((((((((((((((((((..((.....(.....)((..........---------..))...))....)----)))))))))))))))....)))))).. ( -30.70, z-score = -2.13, R) >droSec1.super_0 6501473 99 - 21120651 UCGGUAAGCUGCUUAGCUGCAAGUUGUGAUUAGCACACAGAA--UUUCGCCUAUUUCCC--------CCCGCCCCACCGC---CGUCACAGCUUGCUGCUUUCAAAGCGCGU .......((.(((((((.((((((((((((..((.....(..--...)((.........--------...))......))---.)))))))))))).)))....)))))).. ( -27.20, z-score = -1.53, R) >droYak2.chr3R 11380840 98 + 28832112 UCUGUAAGCUGCUUAGCUGCAAGUUGUGAUUAGCACACAGAA--UUUCGCCUACUUUC--------GCCUUUCCUGCCCC----UUCACAGCUUGCGGCUUUCAAAGCGCGU .......((.(((((((((((((((((((...(((....(((--....((........--------))..))).)))...----.)))))))))))))))....)))))).. ( -32.90, z-score = -2.98, R) >droEre2.scaffold_4770 3471660 98 - 17746568 UCUGUAAGCUGCUUAGCUGCAAGUUGUGAUUAGCACACAGAA--UUUCGCCUAUUUUC--------GCCUUCCUUGCCCC----UUCACAGCUUGCGGCUUUCAAAGCGCGU .......((.(((((((((((((((((((...(((....(((--....((........--------)).)))..)))...----.)))))))))))))))....)))))).. ( -32.20, z-score = -2.88, R) >droAna3.scaffold_13340 13789585 111 + 23697760 UGUAUAAGCAGCUUAGCUGCAACUUGUGAUUAGCACACAAAAG-UCUCACCCCUUUUCCAGCCGCAGCCACCACCACAUCCACAUCCACAGCCAGUGGCUUUCCAAGCGCGU .......((.((((.(((((....((((.....)))).(((((-........)))))......))))).................((((.....))))......)))))).. ( -22.00, z-score = -0.77, R) >dp4.Unknown_group_353 10542 84 + 17550 UUUGUAAGCUGCUUAGCUGCAACUUGUGAUUAGCACACACACCAUGCCACACGCCCCC---------ACGCCACAGAAGC-----UUGCGGCUUGCGU-------------- ...(((((((((..(((((((...((((.........))))...))).....((....---------..))......)))-----).)))))))))..-------------- ( -24.00, z-score = -0.76, R) >consensus UCUGUAAGCUGCUUAGCUGCAAGUUGUGAUUAGCACACAGAA__UUUCGCCUAUUUCC_________CCCCCCCCGCACC____UUCACAGCUUGCGGCUUUCAAAGCGCGU .......((.(((((((((((((((((((........................................................)))))))))))))))....)))))).. (-18.87 = -20.53 + 1.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:42 2011