
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,339,566 – 7,339,657 |
| Length | 91 |
| Max. P | 0.963849 |

| Location | 7,339,566 – 7,339,657 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 71.31 |
| Shannon entropy | 0.49063 |
| G+C content | 0.48802 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -10.52 |
| Energy contribution | -12.68 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
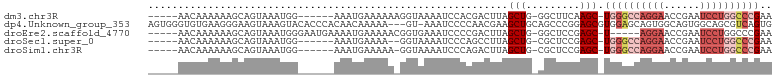
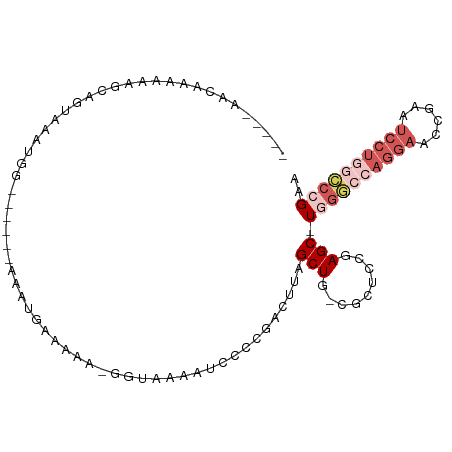
>dm3.chr3R 7339566 91 + 27905053 -----AACAAAAAAGCAGUAAAUGG------AAAUGAAAAAAGGUAAAAUCCACGACUUAGCUG-GGCUUCAAGC-UGGGCCAGGAACCGAAUCCUGGCCCGAA -----........((((((...(((------(.................))))..)))..))).-(((.....))-)(((((((((......)))))))))... ( -27.43, z-score = -2.34, R) >dp4.Unknown_group_353 5487 100 + 17550 AGUGGGUGUGAAGGGAAGUAAAGUACACCCACAACAAAAA---GU-AAAUCCCCAACGAAGCUGCAGCCCGGAGCGUGGAGCAGUGGCAGUGGCAGCGUCAGUG .(((((((((.............)))))))))........---..-...........((.(((((.((((...((.....)).).)))....))))).)).... ( -30.52, z-score = -0.95, R) >droEre2.scaffold_4770 3465877 92 - 17746568 -----AACAAAAAAGCAGUAAAUGGGAAUGAAAAUGAAAAACGGUGAAAUCCCCGACUUAGCUG-GGCUCCGAGC-U-----AGGAACCGAAUCCUGGCCCGAA -----.................((((...((..........(((.(....).))).(((((((.-(....).)))-)-----))).......))....)))).. ( -20.20, z-score = -0.26, R) >droSec1.super_0 6495780 89 - 21120651 -----AACAAAAAAGCAGUAAAUGG------AAAUGAAAA--GGUAAAAUCCCAGCCUUAGCUG-CGCUCCGAGC-UGGGCCAGGAACCGAAUCCUGGCCCGAA -----.........(((((..((..------..))...((--(((.........))))).))))-)((.....))-((((((((((......)))))))))).. ( -31.10, z-score = -3.18, R) >droSim1.chr3R 13463339 90 - 27517382 -----AACAAAAAAGCAGUAAAUGG------AAAUGAAAAA-GGUAAAAUCCCAGACUUAGCUG-CGCUCCGAGC-UGGGCCAGGAACCGAAUCCUGGCCCGAA -----.........(((((...(((------..........-.........)))......))))-)((.....))-((((((((((......)))))))))).. ( -28.41, z-score = -2.81, R) >consensus _____AACAAAAAAGCAGUAAAUGG______AAAUGAAAAA_GGUAAAAUCCCCGACUUAGCUG_CGCUCCGAGC_UGGGCCAGGAACCGAAUCCUGGCCCGAA ............................................................((....))........((((((((((......)))))))))).. (-10.52 = -12.68 + 2.16)
| Location | 7,339,566 – 7,339,657 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 71.31 |
| Shannon entropy | 0.49063 |
| G+C content | 0.48802 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -14.08 |
| Energy contribution | -16.60 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.963849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
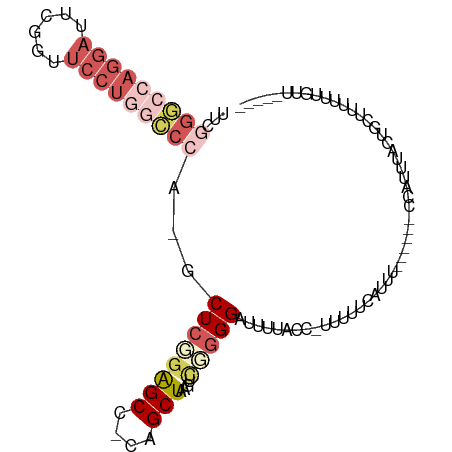
>dm3.chr3R 7339566 91 - 27905053 UUCGGGCCAGGAUUCGGUUCCUGGCCCA-GCUUGAAGCC-CAGCUAAGUCGUGGAUUUUACCUUUUUUCAUUU------CCAUUUACUGCUUUUUUGUU----- ...(((((((((......)))))))))(-((..(((((.-......(((.(((((.................)------))))..))))))))...)))----- ( -28.64, z-score = -2.88, R) >dp4.Unknown_group_353 5487 100 - 17550 CACUGACGCUGCCACUGCCACUGCUCCACGCUCCGGGCUGCAGCUUCGUUGGGGAUUU-AC---UUUUUGUUGUGGGUGUACUUUACUUCCCUUCACACCCACU ((.(((.(((((....(((...((.....))....))).))))).))).)).......-..---........((((((((...............)))))))). ( -29.76, z-score = -1.63, R) >droEre2.scaffold_4770 3465877 92 + 17746568 UUCGGGCCAGGAUUCGGUUCCU-----A-GCUCGGAGCC-CAGCUAAGUCGGGGAUUUCACCGUUUUUCAUUUUCAUUCCCAUUUACUGCUUUUUUGUU----- (((((((.((((......))))-----.-)))))))...-(((.(((((.((((((...................))))))))))))))..........----- ( -24.11, z-score = -1.23, R) >droSec1.super_0 6495780 89 + 21120651 UUCGGGCCAGGAUUCGGUUCCUGGCCCA-GCUCGGAGCG-CAGCUAAGGCUGGGAUUUUACC--UUUUCAUUU------CCAUUUACUGCUUUUUUGUU----- ...(((((((((......)))))))))(-((..(((((.-((((....))))(((.......--........)------)).......)))))...)))----- ( -32.76, z-score = -2.74, R) >droSim1.chr3R 13463339 90 + 27517382 UUCGGGCCAGGAUUCGGUUCCUGGCCCA-GCUCGGAGCG-CAGCUAAGUCUGGGAUUUUACC-UUUUUCAUUU------CCAUUUACUGCUUUUUUGUU----- ...(((((((((......))))))))).-((.....))(-(((.((((..(((((.......-........))------))))))))))).........----- ( -31.16, z-score = -2.87, R) >consensus UUCGGGCCAGGAUUCGGUUCCUGGCCCA_GCUCGGAGCC_CAGCUAAGUCGGGGAUUUUACC_UUUUUCAUUU______CCAUUUACUGCUUUUUUGUU_____ ...(((((((((......)))))))))...((((((((....)))....))))).................................................. (-14.08 = -16.60 + 2.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:39 2011