
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,334,312 – 7,334,409 |
| Length | 97 |
| Max. P | 0.789064 |

| Location | 7,334,312 – 7,334,409 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 67.46 |
| Shannon entropy | 0.64729 |
| G+C content | 0.41883 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -9.08 |
| Energy contribution | -8.15 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.789064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
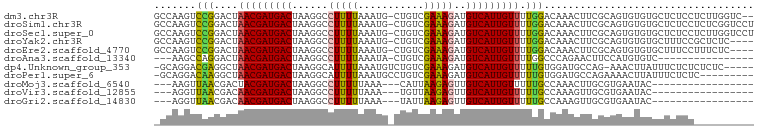
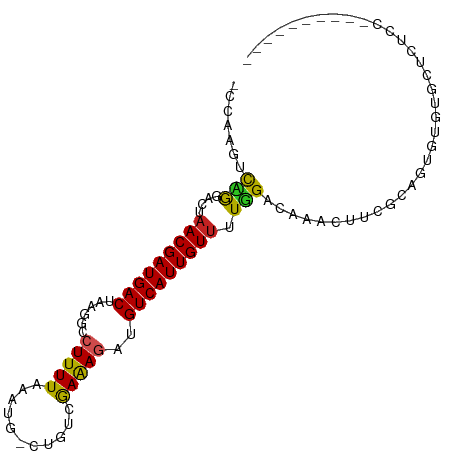
>dm3.chr3R 7334312 97 - 27905053 GCCAAGUCCGGACUAACGAUGACUAAGGCCUUUUAAAUG-CUGUCGAAAGAUGUCAUUGUUUUGGACAAACUUCGCAGUGUGUGCUCUCCUCUUGGUC-- ((((((((((((...((((((((...(((.........)-))(((....)))))))))))))))))).......(((.....))).......))))).-- ( -28.10, z-score = -2.08, R) >droSim1.chr3R 13456558 99 + 27517382 GCCAAGUCCGGACUAACGAUGACUAAGGCCUUUUAAAUG-CUGUCGAAAGAUGUCAUUGUUUUGGACAAACUUCGCAGUGUGUGCUCUCCUCUCGGUCCU (((..(((((((...((((((((...(((.........)-))(((....)))))))))))))))))).......(((.....))).........)))... ( -26.70, z-score = -1.43, R) >droSec1.super_0 6490587 99 + 21120651 GCCAAGUCCGGACUAACGAUGACUAAGGCCUUUUAAAUG-CUGUCGAAAGAUGUCAUUGUUUUGGACAAACUUCGCAGUGUGUGCUCUCCUCUUGGUCCU ((((((((((((...((((((((...(((.........)-))(((....)))))))))))))))))).......(((.....))).......)))))... ( -28.10, z-score = -1.99, R) >droYak2.chr3R 11369629 95 - 28832112 GCCAAGUCCGGACUAACGAUGACUAAGGCCUUUUAAAUG-CUGUCGAAAGAUGUCAUUGUUUUGGACAAACUUCGCAGUGUGUGCUUUCCGCUCUC---- ((...(((((((...((((((((...(((.........)-))(((....)))))))))))))))))).......(((.....))).....))....---- ( -25.00, z-score = -1.32, R) >droEre2.scaffold_4770 3460691 95 + 17746568 GCCAAGUCCGGACUAACGAUGACUAAGGCCUUUUAAAUG-CUGUCGAAAGAUGUCAUUGUUUUGGACAAACUUCGCAGUGUGUGCUUUCCUUUCUC---- .....(((((((...((((((((...(((.........)-))(((....)))))))))))))))))).......(((.....)))...........---- ( -23.90, z-score = -1.31, R) >droAna3.scaffold_13340 13773595 80 - 23697760 ---AAGCCAGGACUAACGAUGACUAAGGCCUUUUAAAUA-CUGUCGAAAGAUGUCAUUGUUUUGGCCCAGAACUUCCAUGUGUC---------------- ---..((((((((....((((((..((............-))(((....)))))))))))))))))..................---------------- ( -18.10, z-score = -0.99, R) >dp4.Unknown_group_353 866 93 - 17550 -GCAGGACGAGGCUAACGAUGACUAAGGCAUUUUAAAUGUCUGUCGAAAGAUGUCAUUGUUUUUGUGGAUGCCAG-AAACUUAUUUCUCUCUCUC----- -...(((.((((..(((((((((..((((((.....))))))(((....))))))))))))(((.(((...))).-)))......)))))))...----- ( -24.70, z-score = -1.66, R) >droPer1.super_6 957686 90 - 6141320 -GCAGGACAAGGCUAACGAUGACUAAGGCAUUUUAAAUGCCUGUCGAAAGAUGUCAUUGUUUUUGUGGAUGCCAGAAAACUUAUUUCUCUC--------- -(((..(((((...(((((((((..((((((.....))))))(((....)))))))))))))))))...))).(((((.....)))))...--------- ( -27.40, z-score = -3.15, R) >droMoj3.scaffold_6540 24831710 77 - 34148556 ---AAGUUAACGACUACGAUGACUAAGGCCUUUUUAAA---CAUUAAGAGUUGUCAUUGUUUUUGCCAAACUUGCGUGAAUAC----------------- ---(((((..(((..((((((((......(((((....---....)))))..))))))))..)))...)))))..........----------------- ( -13.60, z-score = -0.76, R) >droVir3.scaffold_12855 9830168 77 - 10161210 ---AGGUUAACGACAACGAUGACUAAGGCCUUUUUAAA---UGUUAAGAGUUGUCAUUGUUUUUGCCAAAGUUGCGUGAAUAC----------------- ---.(((....(((((.((..(((..(((.((....))---.)))...)))..)).)))))...)))................----------------- ( -13.80, z-score = -0.10, R) >droGri2.scaffold_14830 2172912 77 - 6267026 ---AGGUUAACGACAACGAUGACUAAGGCCUUUUUAAA---UAUUAAGAGUUGUCAUUGUUUUUGCCAAAGUUGCGUGAAUAC----------------- ---.(((....((.(((((((((......(((((....---....)))))..))))))))).)))))................----------------- ( -12.80, z-score = -0.10, R) >consensus _CCAAGUCAGGACUAACGAUGACUAAGGCCUUUUAAAUG_CUGUCGAAAGAUGUCAUUGUUUUGGACAAACUUCGCAGUGUGUGCUCUCC__________ .......(((....(((((((((......(((((...........)))))..))))))))).)))................................... ( -9.08 = -8.15 + -0.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:37 2011