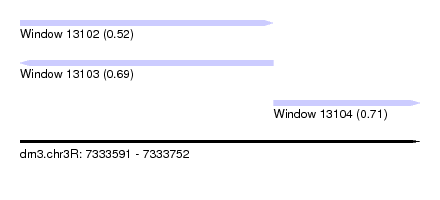
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,333,591 – 7,333,752 |
| Length | 161 |
| Max. P | 0.708906 |

| Location | 7,333,591 – 7,333,693 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 65.31 |
| Shannon entropy | 0.63130 |
| G+C content | 0.40176 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -9.46 |
| Energy contribution | -12.30 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.515314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
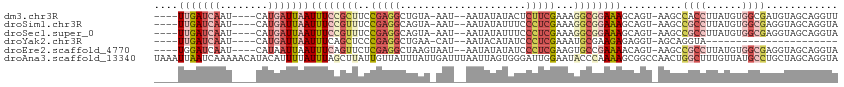
>dm3.chr3R 7333591 102 + 27905053 ----UUGAUCAAU----CAUGAUUAAUUUCCGCUUCCGAGGCUGUA-AAU--AAUAUAUACUCUUCGAAAGGCGGAAAGCAGU-AAGCCACCUUAUGUGGCGAUGUAGCAGGUU ----(((((((..----..)))))))(((((((((.(((((..(((-...--......))).)))))..)))))))))((..(-(.(((((.....)))))..))..))..... ( -30.60, z-score = -2.60, R) >droSim1.chr3R 13455828 102 - 27517382 ----UUGAUCAAU----CAUGAUUAAUUUCCGUUUCCGAGGCAGUA-AAU--AAUAUAUUUCCCUCGAAAGGCGGAAAGCAGU-AAGCCGCCUUAUGUGGCGAGGUAGCAGGUA ----(((((((..----..)))))))(((((((((.(((((.((((-...--....))))..)))))..)))))))))((..(-..(((((.....)))))...)..))..... ( -29.50, z-score = -2.15, R) >droSec1.super_0 6489857 102 - 21120651 ----UUGAUCAAU----CAUGAUUAAUUUCCGUUUCCGAGGCAGUA-AAU--AAUAUAUUUCCCUCGAAAGGCGGAAAGCAGU-AAGCCGCCUUAUGUGGCGAGGUAGCAGGUA ----(((((((..----..)))))))(((((((((.(((((.((((-...--....))))..)))))..)))))))))((..(-..(((((.....)))))...)..))..... ( -29.50, z-score = -2.15, R) >droYak2.chr3R 11368891 80 + 28832112 ----UUGAUCAAU----CAUGAUUAAUUUCAGCUCCCGAGGCUGAA-CAU--AAUACAUAUCCCUCGAAAUGCGAAGAGAGGU-AGCAGGUA---------------------- ----(((((((..----..))))))).(((((((.....)))))))-...--..(((.((((.(((..........))).)))-)....)))---------------------- ( -17.00, z-score = -0.73, R) >droEre2.scaffold_4770 3459967 103 - 17746568 ----UGGAUCAAU----CAUAAUUAAUUUCAGUUCUCGAGGCUAAGUAAU--AAUAUAUAUCCCUCGAAGUGCCGAAAACAGU-AAGCCGCCUUAUGUGGCGAGGUAGCAGGUA ----...(((...----.........((((.((.(((((((.((.(((..--..))).))..))))).)).)).))))...((-.(.(((((......)))).).).)).))). ( -20.70, z-score = 0.10, R) >droAna3.scaffold_13340 13772769 114 + 23697760 UAAAUUAAUCAAAAACAUACAUUUUAUUUAGCUUAUUGUUAUUUAUUGAUUUAAUUAGUGGGAUUGGAAUACCCAAAAGCGGCCAACUGGCUUUGUUAUGCCUGCUAGCAGGUA .............((((.............((((.......(((((((((...))))))))).((((.....))))))))((((....)))).)))).((((((....)))))) ( -22.70, z-score = -0.11, R) >consensus ____UUGAUCAAU____CAUGAUUAAUUUCCGUUUCCGAGGCUGAA_AAU__AAUAUAUAUCCCUCGAAAGGCGGAAAGCAGU_AAGCCGCCUUAUGUGGCGAGGUAGCAGGUA ....(((((((........)))))))((((((((..(((((.....................)))))...))))))))..........((((......))))............ ( -9.46 = -12.30 + 2.84)
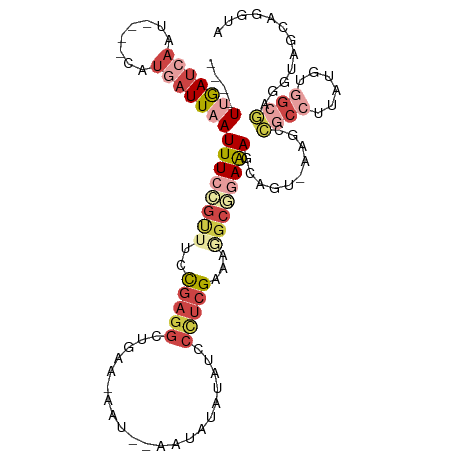
| Location | 7,333,591 – 7,333,693 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 65.31 |
| Shannon entropy | 0.63130 |
| G+C content | 0.40176 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -9.81 |
| Energy contribution | -11.28 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7333591 102 - 27905053 AACCUGCUACAUCGCCACAUAAGGUGGCUU-ACUGCUUUCCGCCUUUCGAAGAGUAUAUAUU--AUU-UACAGCCUCGGAAGCGGAAAUUAAUCAUG----AUUGAUCAA---- .....((......(((((.....)))))..-...))(((((((.((((((.(.(((......--...-)))..).)))))))))))))...((((..----..))))...---- ( -25.30, z-score = -1.84, R) >droSim1.chr3R 13455828 102 + 27517382 UACCUGCUACCUCGCCACAUAAGGCGGCUU-ACUGCUUUCCGCCUUUCGAGGGAAAUAUAUU--AUU-UACUGCCUCGGAAACGGAAAUUAAUCAUG----AUUGAUCAA---- .....((....(((((......)))))...-...))((((((..((((((((.(((((...)--)))-)....)))))))).))))))...((((..----..))))...---- ( -25.40, z-score = -2.09, R) >droSec1.super_0 6489857 102 + 21120651 UACCUGCUACCUCGCCACAUAAGGCGGCUU-ACUGCUUUCCGCCUUUCGAGGGAAAUAUAUU--AUU-UACUGCCUCGGAAACGGAAAUUAAUCAUG----AUUGAUCAA---- .....((....(((((......)))))...-...))((((((..((((((((.(((((...)--)))-)....)))))))).))))))...((((..----..))))...---- ( -25.40, z-score = -2.09, R) >droYak2.chr3R 11368891 80 - 28832112 ----------------------UACCUGCU-ACCUCUCUUCGCAUUUCGAGGGAUAUGUAUU--AUG-UUCAGCCUCGGGAGCUGAAAUUAAUCAUG----AUUGAUCAA---- ----------------------........-...((.(((((.....)))))))((((.(((--(..-(((((((....).))))))..))))))))----.........---- ( -15.60, z-score = 0.21, R) >droEre2.scaffold_4770 3459967 103 + 17746568 UACCUGCUACCUCGCCACAUAAGGCGGCUU-ACUGUUUUCGGCACUUCGAGGGAUAUAUAUU--AUUACUUAGCCUCGAGAACUGAAAUUAAUUAUG----AUUGAUCCA---- ...........(((((......)))))...-.....((((((...(((((((..((..((..--..))..)).)))))))..)))))).........----.........---- ( -20.10, z-score = -0.46, R) >droAna3.scaffold_13340 13772769 114 - 23697760 UACCUGCUAGCAGGCAUAACAAAGCCAGUUGGCCGCUUUUGGGUAUUCCAAUCCCACUAAUUAAAUCAAUAAAUAACAAUAAGCUAAAUAAAAUGUAUGUUUUUGAUUAAUUUA ((((((..(((.(((.((((.......))))))))))..))))))..................((((((.((((((((...............))).)))))))))))...... ( -19.86, z-score = -0.20, R) >consensus UACCUGCUACCUCGCCACAUAAGGCGGCUU_ACUGCUUUCCGCCUUUCGAGGGAUAUAUAUU__AUU_UACAGCCUCGGAAACGGAAAUUAAUCAUG____AUUGAUCAA____ ............((((......))))..........((((((...(((((((.....................)))))))..)))))).......................... ( -9.81 = -11.28 + 1.48)

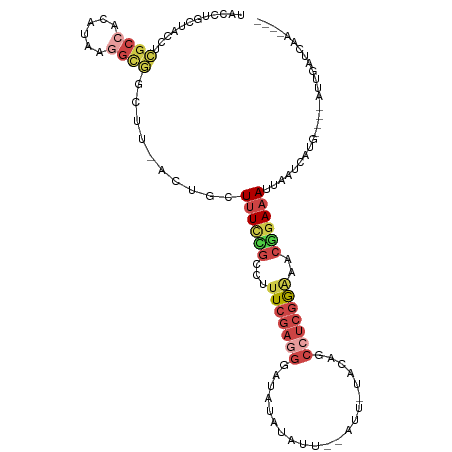
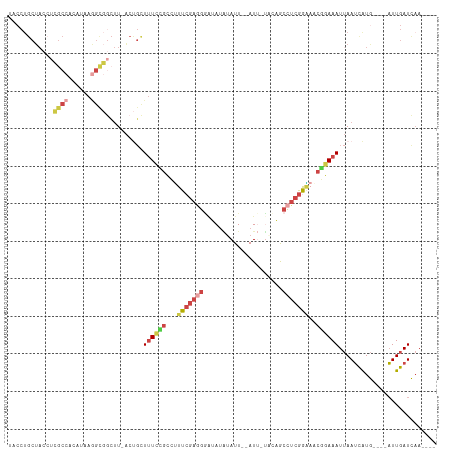
| Location | 7,333,693 – 7,333,752 |
|---|---|
| Length | 59 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | forward |
| Mean pairwise identity | 87.77 |
| Shannon entropy | 0.21522 |
| G+C content | 0.49825 |
| Mean single sequence MFE | -12.90 |
| Consensus MFE | -11.92 |
| Energy contribution | -11.52 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708906 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 7333693 59 + 27905053 GUAAAAAGUGCGUAUACGCCCUGUGUUCGCCUUGUCUGCCGUGGUGUUGGUAUUCUUUC-- .....(((.(((.((((.....)))).))))))(..(((((......)))))..)....-- ( -12.10, z-score = -1.38, R) >droEre2.scaffold_4770 3460070 56 - 17746568 -GGAAAAGUGCGUAUACGCCCUGUGUUCGUCUUUUCUGCCGUGGUGUUGGUAUUGGU---- -(((((((.(((.((((.....)))).))))))))))((((......))))......---- ( -14.40, z-score = -1.40, R) >droYak2.chr3R 11368971 60 + 28832112 -GGGAAAGUGCGUAUACGCCCUGUGUUCGCCUUGUCUGCCGUGGUGUUGGUAUUAGUAUUG -.((((((.(((.((((.....)))).)))))).)))((((......)))).......... ( -12.80, z-score = 0.32, R) >droSec1.super_0 6489959 59 - 21120651 GUAAAAAGUGCGUAUACGCCCUGUGUUCGCCUUGUCUGCCAUGGUGUUGGUAUUCCUUC-- .....(((.(((.((((.....)))).))))))...(((((......))))).......-- ( -12.60, z-score = -1.57, R) >droSim1.chr3R 13455930 59 - 27517382 GUAAAAAGUGCGUAUACGCCCUGUGUUCGCCUUGUCUGCCAUGGUGUUGGUAUUCCUUC-- .....(((.(((.((((.....)))).))))))...(((((......))))).......-- ( -12.60, z-score = -1.57, R) >consensus GUAAAAAGUGCGUAUACGCCCUGUGUUCGCCUUGUCUGCCGUGGUGUUGGUAUUCCUUC__ .....(((.(((.((((.....)))).))))))...(((((......)))))......... (-11.92 = -11.52 + -0.40)

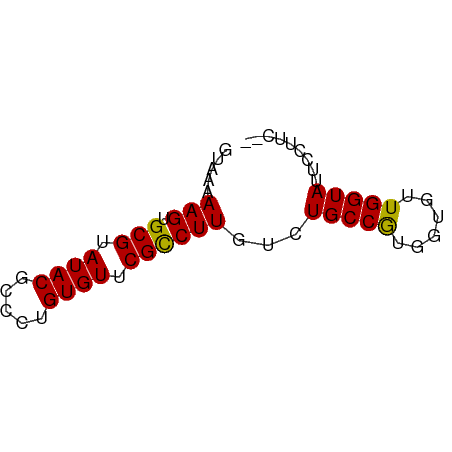
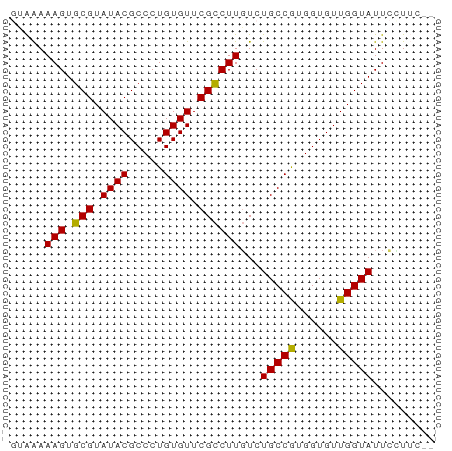
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:36 2011