
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,303,051 – 7,303,184 |
| Length | 133 |
| Max. P | 0.888470 |

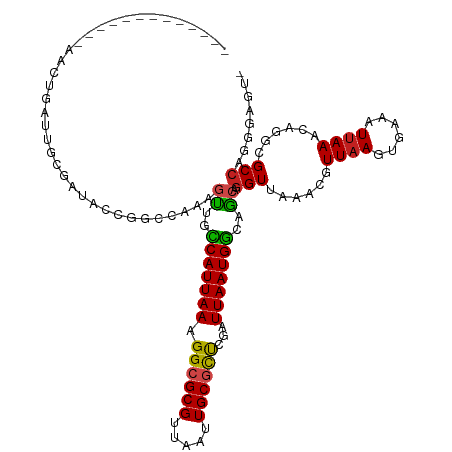
| Location | 7,303,051 – 7,303,157 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.53 |
| Shannon entropy | 0.43607 |
| G+C content | 0.46595 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -17.17 |
| Energy contribution | -16.70 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.574933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7303051 106 - 27905053 -------------AACUGAUUGCGAUGCCGGCCAAAGUUGCCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCAGGUUAAACGUUAAGUGAAAUUAAACAGGCGCCAGGGAGU- -------------.(((..(((.(.((((((((...(((((((((((.((((((.....))))))...))))))))))).)))).........((........)).))))))))..)))- ( -35.80, z-score = -2.03, R) >droSim1.chr3R 13427765 106 + 27517382 -------------AACUGAUUGCGAUGCCGGCCAAAGUUGCCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCAGGUUAAACGUUAAGUGAAAUUAAACAGGCGCCAGGGAGU- -------------.(((..(((.(.((((((((...(((((((((((.((((((.....))))))...))))))))))).)))).........((........)).))))))))..)))- ( -35.80, z-score = -2.03, R) >droSec1.super_0 6459663 106 + 21120651 -------------AACUGAUUGCGAUGCCGGCCAAAGUUGCCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCAGGUUAAACGUUAAGUGAAAUUAAACAGGCGCCAGGGAGU- -------------.(((..(((.(.((((((((...(((((((((((.((((((.....))))))...))))))))))).)))).........((........)).))))))))..)))- ( -35.80, z-score = -2.03, R) >droYak2.chr3R 11337750 106 - 28832112 -------------AACUGAUUGCGAUGCCGGCCAAAGUUGCCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCAGGUUAAACGUUAAGUGAAAUUAAACAGGCGCCAGGGAGU- -------------.(((..(((.(.((((((((...(((((((((((.((((((.....))))))...))))))))))).)))).........((........)).))))))))..)))- ( -35.80, z-score = -2.03, R) >droEre2.scaffold_4770 3429336 106 + 17746568 -------------AACUGAUUGCGAUGCCGGCCAAAGUUGCCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCAGGUUAAACGUUAAGUGAAAUUAAACAGGCGCCAGGGAGU- -------------.(((..(((.(.((((((((...(((((((((((.((((((.....))))))...))))))))))).)))).........((........)).))))))))..)))- ( -35.80, z-score = -2.03, R) >droAna3.scaffold_13340 13737254 106 - 23697760 -------------AACUGAUUGCGAAACUGGCCAGAGUUUCCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCAGGUUAAACGUUAAGUGAAAUUAAACAGGCGCCAGGGAGU- -------------..(((...(((...((((((...(((.(((((((.((((((.....))))))...))))))).))).)))...((.....)).........))).)))))).....- ( -27.60, z-score = -0.25, R) >dp4.chr2 25597699 106 - 30794189 ------------UGAAGCACUGGGCCAGAGGCAGUGCUGGCCAUUAAAGGCGCGUUAAUUGCACUCGAUUAAUGGCAGCAGGUUAAACGUUAAGUGAAAUUAAACAGGCGCCAGGGAA-- ------------.......(((((((...(((..(((((.(((((((.((.(((.....))).))...)))))))))))).))).........((........)).))).))))....-- ( -32.20, z-score = -0.92, R) >droPer1.super_6 926064 106 - 6141320 ------------AGAAGCACUGGGCCAGAGGCAGUGCUGGCCAUUAAAGGCGCGUUAAUUGCACUCGAUUAAUGGCAGCAGGUUAAACGUUAAGUGAAAUUAAACAGGCGCCAGGGAA-- ------------.......(((((((...(((..(((((.(((((((.((.(((.....))).))...)))))))))))).))).........((........)).))).))))....-- ( -32.20, z-score = -1.05, R) >droWil1.scaffold_181089 8191642 119 + 12369635 AACAAAAAUUGGACAUGAACACGGUCAGAGA-GUUGGCGCCCAUUAAAGGCGCGUUAAUUGCGCUCAAUUAAUGGCAGCAGGUUAAACGUUAAGUAAAAUUAAACAGACGCCAGGGACGG .....................(.(((.....-.((((((((((((((.((((((.....))))))...)))))))..))..(((.....((((......))))...)))))))).))).) ( -25.70, z-score = 0.01, R) >droVir3.scaffold_12855 9787712 93 - 10161210 --------------------------AUGGCCGGAGGGCAUCAUUAAAGGCGCGUUAAUUGCGCCCAAUUAAUGACAUCAGGUUAAACGUUAAGUGAAAUGAAACAGACGCCUGGGAAU- --------------------------...(((....))).(((((((.((((((.....))))))...)))))))..(((((.....((((..((........)).)))))))))....- ( -25.70, z-score = -1.39, R) >droMoj3.scaffold_6540 24792704 93 - 34148556 --------------------------ACGACCGGAGGGCGUCAUUAAAGGCGCGUUAAUUGCGCUCAAUUAAUGACAUCAGGUUAAACGUUAAGUGAAAUUAAACAGACGCCCGGGACU- --------------------------....((...(((((((.......(((((.....)))))(((.((((((.............)))))).))).........))))))).))...- ( -28.32, z-score = -2.35, R) >droGri2.scaffold_14830 2132601 91 - 6267026 --------------------------AUGACCAGAGGGCGUCAUUAAAGGCGCGUUAAUUGCGUCCAAUUAAUGGCAUCAGGUUAAACGUUAAGUGAAAUUAAACAGAUGCGUGGGG--- --------------------------.(((((.((....((((((((.((((((.....))))))...)))))))).)).))))).((((...((........))....))))....--- ( -23.30, z-score = -1.39, R) >consensus _____________AACUGAUUGCGAUACCGGCCAAAGUUGCCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCAGGUUAAACGUUAAGUGAAAUUAAACAGGCGCCAGGGAGU_ ....................................((..(((((((.((((((.....))))))...)))))))..)).(((......((((......))))......)))........ (-17.17 = -16.70 + -0.47)


| Location | 7,303,090 – 7,303,184 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 65.54 |
| Shannon entropy | 0.64945 |
| G+C content | 0.48366 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -10.32 |
| Energy contribution | -10.40 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7303090 94 + 27905053 UGCUGCCAUUAAUCGAGCGCAAUUAACGCGCCUUUAAUGG----CAA--CUUUGGCCGGCAUCGCAAUCAGUUGGCUCACACACA-CACACACUUACUGGC---- .(((((((((((..(.((((.......))))).)))))))----)..--....(((((((..........)))))))........-............)))---- ( -25.80, z-score = -1.61, R) >droYak2.chr3R 11337789 98 + 28832112 UGCUGCCAUUAAUCGAGCGCAAUUAACGCGCCUUUAAUGG----CAA--CUUUGGCCGGCAUCGCAAUCAGUUGGCCAACACACA-CACACACGCAUUGACUGGC ((((((((((((..(.((((.......))))).)))))))----)).--..(((((((((..........)))))))))......-.......)))......... ( -32.40, z-score = -2.65, R) >droEre2.scaffold_4770 3429375 92 - 17746568 UGCUGCCAUUAAUCGAGCGCAAUUAACGCGCCUUUAAUGG----CAA--CUUUGGCCGGCAUCGCAAUCAGUUGGCUCACACACA-CACAUAG--ACUGGC---- .(((((((((((..(.((((.......))))).)))))))----)..--....(((((((..........)))))))........-.......--...)))---- ( -25.80, z-score = -1.18, R) >droAna3.scaffold_13340 13737293 94 + 23697760 UGCUGCCAUUAAUCGAGCGCAAUUAACGCGCCUUUAAUGG----AAA--CUCUGGCCAGUUUCGCAAUCAGUUUACCCUCACACA-CACACUACCACUCAC---- (((...((((((..(.((((.......))))).))))))(----(((--((......)))))))))...................-...............---- ( -17.30, z-score = -1.40, R) >droWil1.scaffold_181089 8191682 102 - 12369635 UGCUGCCAUUAAUUGAGCGCAAUUAACGCGCCUUUAAUGGGCGCCAACUCUCUGACCGUGUUCAUGUCCAAUUUUUGU-UGUAUUGUGUACACACACACACAC-- (((.((((.....)).)))))......((((((.....)))))).............((((.((((..((((....))-))...)))).))))..........-- ( -23.70, z-score = -0.48, R) >droVir3.scaffold_12855 9787751 76 + 10161210 UGAUGUCAUUAAUUGGGCGCAAUUAACGCGCCUUUAAUGA-UGCC----CUCCGGCCAUGUCCGGCGUGUAU----AUAUGUAUA-------------------- ....((((((((..((((((.......)))))))))))))-)((.----(.((((......)))).).))..----.........-------------------- ( -23.40, z-score = -1.28, R) >droGri2.scaffold_14830 2132638 86 + 6267026 UGAUGCCAUUAAUUGGACGCAAUUAACGCGCCUUUAAUGA-CGCC----CUCUGGUCAUGUCCAGCAUAAACUUUGAUAUGCACAGGGUAC-------------- ......((((((..((.(((.......))))).)))))).-.(((----((.(((......)))(((((........)))))..)))))..-------------- ( -23.40, z-score = -1.74, R) >consensus UGCUGCCAUUAAUCGAGCGCAAUUAACGCGCCUUUAAUGG____CAA__CUCUGGCCGGCUUCGCAAUCAGUUGGCACACACACA_CACACAC__ACUGAC____ .....(((((((....((((.......))))..)))))))................................................................. (-10.32 = -10.40 + 0.08)



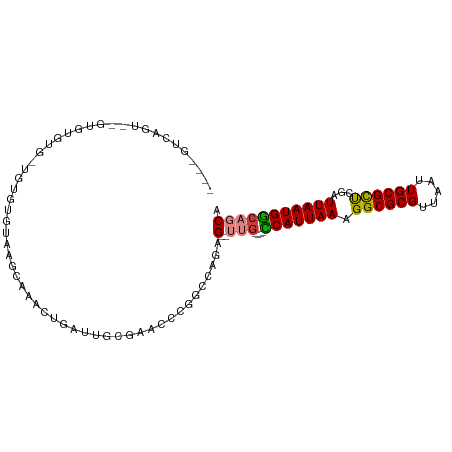
| Location | 7,303,090 – 7,303,184 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 65.54 |
| Shannon entropy | 0.64945 |
| G+C content | 0.48366 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -14.45 |
| Energy contribution | -13.80 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7303090 94 - 27905053 ----GCCAGUAAGUGUGUG-UGUGUGUGAGCCAACUGAUUGCGAUGCCGGCCAAAG--UUG----CCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCA ----(((.(((.(((....-((.((....))))......)))..))).)))....(--(((----(((((((.((((((.....))))))...))))))))))). ( -31.40, z-score = -0.76, R) >droYak2.chr3R 11337789 98 - 28832112 GCCAGUCAAUGCGUGUGUG-UGUGUGUUGGCCAACUGAUUGCGAUGCCGGCCAAAG--UUG----CCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCA (((.(((((((((......-..))))))))).........((...)).)))....(--(((----(((((((.((((((.....))))))...))))))))))). ( -35.40, z-score = -0.77, R) >droEre2.scaffold_4770 3429375 92 + 17746568 ----GCCAGU--CUAUGUG-UGUGUGUGAGCCAACUGAUUGCGAUGCCGGCCAAAG--UUG----CCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCA ----((((((--(....((-.((......))))...))))((...)).)))....(--(((----(((((((.((((((.....))))))...))))))))))). ( -30.40, z-score = -0.64, R) >droAna3.scaffold_13340 13737293 94 - 23697760 ----GUGAGUGGUAGUGUG-UGUGUGAGGGUAAACUGAUUGCGAAACUGGCCAGAG--UUU----CCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCA ----.....(((((((...-((((...((.....))...))))..))).))))..(--((.----(((((((.((((((.....))))))...))))))).))). ( -26.90, z-score = -0.66, R) >droWil1.scaffold_181089 8191682 102 + 12369635 --GUGUGUGUGUGUGUACACAAUACA-ACAAAAAUUGGACAUGAACACGGUCAGAGAGUUGGCGCCCAUUAAAGGCGCGUUAAUUGCGCUCAAUUAAUGGCAGCA --...(((((.(((((...((((...-......)))).))))).)))))(((((....)))))(((((((((.((((((.....))))))...)))))))..)). ( -28.50, z-score = -0.26, R) >droVir3.scaffold_12855 9787751 76 - 10161210 --------------------UAUACAUAU----AUACACGCCGGACAUGGCCGGAG----GGCA-UCAUUAAAGGCGCGUUAAUUGCGCCCAAUUAAUGACAUCA --------------------.........----.....(.((((......)))).)----((..-(((((((.((((((.....))))))...)))))))..)). ( -22.40, z-score = -2.15, R) >droGri2.scaffold_14830 2132638 86 - 6267026 --------------GUACCCUGUGCAUAUCAAAGUUUAUGCUGGACAUGACCAGAG----GGCG-UCAUUAAAGGCGCGUUAAUUGCGUCCAAUUAAUGGCAUCA --------------...((((..(((((........)))))(((......))).))----)).(-(((((((.((((((.....))))))...)))))))).... ( -27.30, z-score = -2.08, R) >consensus ____GUCAGU__GUGUGUG_UGUGUGUAAGCAAACUGAUUGCGAACCCGGCCAGAG__UUG____CCAUUAAAGGCGCGUUAAUUGCGCUCGAUUAAUGGCAGCA .................................................................(((((((.((((((.....))))))...)))))))..... (-14.45 = -13.80 + -0.65)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:30 2011