
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,287,045 – 7,287,110 |
| Length | 65 |
| Max. P | 0.554974 |

| Location | 7,287,045 – 7,287,110 |
|---|---|
| Length | 65 |
| Sequences | 5 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 65.27 |
| Shannon entropy | 0.59538 |
| G+C content | 0.37738 |
| Mean single sequence MFE | -5.94 |
| Consensus MFE | -3.76 |
| Energy contribution | -4.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.35 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7287045 65 - 27905053 CGAGUGCAAUUACUUUUGUUAAUUGUAAUACACACAACC--CCAACCAACCAACCAACGCAGAGACA ....(((((((((....).))))))))............--.......................... ( -5.10, z-score = -0.05, R) >droPer1.super_6 910371 54 - 6141320 CGAUUGCAAUUACUUUUGUUAAUUGUAAUACCCACA-----CAAGCACACAGAUAGACA-------- ..(((((((((((....).)))))))))).......-----..................-------- ( -7.20, z-score = -1.66, R) >dp4.chr2 25582656 54 - 30794189 CGAUUGCAAUUACUUUUGUUAAUUGUAAUACACAUA-----CAAGCACACAGACAGACA-------- ..(((((((((((....).)))))))))).......-----..................-------- ( -7.20, z-score = -1.42, R) >droSec1.super_0 6443987 67 + 21120651 CGAGUGCAAUUACAUUUGUUAAUUGUAAUACACACAACCAACCAAGCAACCAACCGACGCAGAGACA ...(((..((((((.........)))))).)))............((...........))....... ( -5.80, z-score = 0.14, R) >droVir3.scaffold_12855 9742745 57 - 10161210 ------CAAUUACUUUUGCUAAUUGUAAUAGACGCUCGCCAUUAUCUAACAGCUGCACAGAUA---- ------......((.((((.....)))).))..((..((............)).)).......---- ( -4.40, z-score = 1.23, R) >consensus CGAGUGCAAUUACUUUUGUUAAUUGUAAUACACACA__C__CAAACAAACAGACAGACA_A_A____ ....(((((((((....).))))))))........................................ ( -3.76 = -4.16 + 0.40)

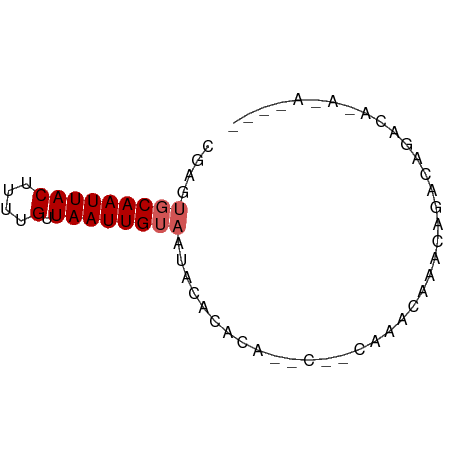
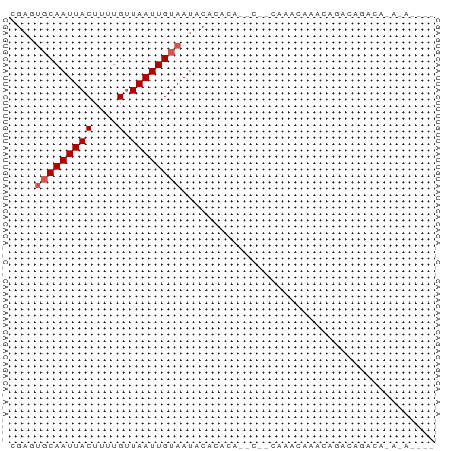
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:26 2011