| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,035,000 – 7,035,094 |
| Length | 94 |
| Max. P | 0.995308 |

| Location | 7,035,000 – 7,035,094 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.85 |
| Shannon entropy | 0.46071 |
| G+C content | 0.30585 |
| Mean single sequence MFE | -18.31 |
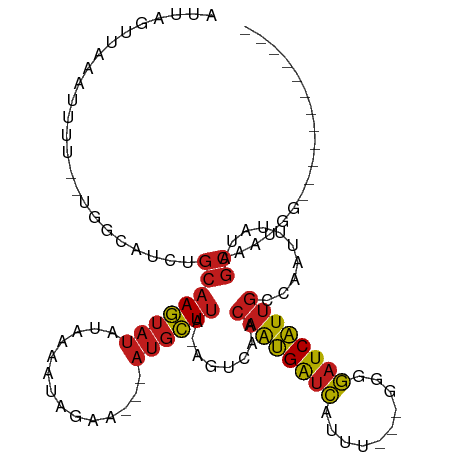
| Consensus MFE | -13.40 |
| Energy contribution | -13.30 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
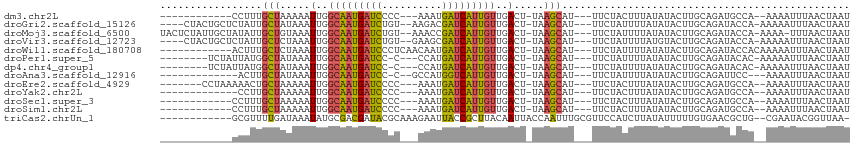
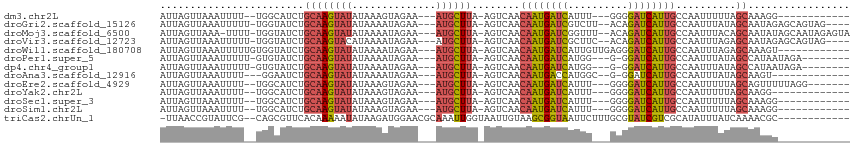
>dm3.chr2L 7035000 94 + 23011544 ------------CCUUUGCUAAAAAUUGGCAAUGAUCCCC---AAAUGAUCAUUGUUGACU-UAAGCAU---UUCUACUUUAUAUACUUGCAGAUGCCA--AAAAUUUAACUAAU ------------....((((.....(..(((((((((...---....)))))))))..)..-..)))).---.................((....))..--.............. ( -16.40, z-score = -2.14, R) >droGri2.scaffold_15126 610435 104 + 8399593 ----CUACUGCUCUAUUGCUAUAAAUUGGCAAUGAUCUGU--AAGACGAUCAUUGUUGACU-UAAGCAU---UUCUAUUUUAUAUACUUGCAGAUACCA-AAAAAUUUAACUAAU ----...((((.....((((.(((.(..(((((((((...--.....)))))))))..).)-)))))).---...(((.....)))...))))......-............... ( -22.30, z-score = -3.32, R) >droMoj3.scaffold_6500 16207925 107 - 32352404 UACUCUAUUGCUAUAUUGCUGUAAAUUGGCAAUGAUCUGU--AAACCGAUCAUUGUUGACU-UAAGCAU---UUCUAUUUUAUAUACUUGCAGAUACCA-AAAA-UUUAACUAAU .......(((((((((((((.(((.(..(((((((((.(.--...).)))))))))..).)-)))))).---.........)))))...))))......-....-.......... ( -20.50, z-score = -2.65, R) >droVir3.scaffold_12723 2924079 104 - 5802038 ----CUACUGCUCUAUUGCUCUAAAUUGGCAAUGAUCUGU--GAAGCGAUCAUUGUUGACU-UAAGCAU---UUCUAUUUUAUGUACUUGCAGAUACCA-AAAAAUUUAACUAAU ----...((((.....((((.(((.(..(((((((((...--.....)))))))))..).)-)))))).---...(((.....)))...))))......-............... ( -22.60, z-score = -2.75, R) >droWil1.scaffold_180708 5106609 99 + 12563649 ------------ACUUUGCUCUAAAUUGGCAAUGAUCCCUCAACAAUGAUCAUUGUUGACU-UAAGCAU---UUCUAUUUUAUAUACUUGCAGAUACCACAAAAAUUUAACUAAU ------------....((((.(((.(..(((((((((..........)))))))))..).)-)))))).---........................................... ( -17.30, z-score = -2.84, R) >droPer1.super_5 1603111 98 + 6813705 --------UCUAUUAUGGCUAUAAAUUGGCAAUGAUCC-C---CCAUGAUCAUUGUUGACU-UAAGCAU---UUCUAUUUUAUAUACUUGCAGAUACAC-AAAAAUUUAACUAAU --------.........(((.(((.(..(((((((((.-.---....)))))))))..).)-)))))..---...........................-............... ( -17.70, z-score = -2.09, R) >dp4.chr4_group1 1668128 98 - 5278887 --------UCUAUUAUGGCUAUAAAUUGGCAAUGAUCC-C---CCAUGAUCAUUGUUGACU-UAAGCAU---UUCUAUUUUAUAUACUUGCAGAUACAC-AAAAAUUUAACUAAU --------.........(((.(((.(..(((((((((.-.---....)))))))))..).)-)))))..---...........................-............... ( -17.70, z-score = -2.09, R) >droAna3.scaffold_12916 106413 92 + 16180835 -------------ACUUGCUAUAAAUUGGCAAUGAUCC-C--GCCAUGGUCAUUGUUGACU-UAAGCAU---UUCUAUUUUAUAUACUUGCAGAUUCC---AAAAUUUAACUAAU -------------...((((.(((.(..(((((((((.-.--.....)))))))))..).)-)))))).---..........................---.............. ( -17.30, z-score = -1.82, R) >droEre2.scaffold_4929 15945551 99 + 26641161 -------CCUAAAAACUGCUAAAAAUUGGCAAUGAUCCCC---AAAUGAUCAUUGUUGACU-UAAGCAU---UUCUACUUUAUAUACUUGCAGAUGCCA--AAAAUUUAACUAAU -------........((((......(..(((((((((...---....)))))))))..)..-.(((...---.....))).........))))......--.............. ( -16.80, z-score = -2.64, R) >droYak2.chr2L 16445868 93 - 22324452 -------------CCUUGCUAAAAAUUGGCAAUGAUCCCC---AAAUGAUCAUUGUUGACU-UAAGCAU---UUCUACUUUAUAUACUUGCAGAUGCCA--AAAAUUUAACUAAU -------------...((((.....(..(((((((((...---....)))))))))..)..-..)))).---.................((....))..--.............. ( -16.40, z-score = -2.22, R) >droSec1.super_3 2559980 94 + 7220098 ------------CCUUUGCUAAAAAUUGGCAAUGAUCCCC---AAAUGAUCAUUGUUGACU-UAAGCAU---UUCUACUUUAUAUACUUGCAGAUGCCA--AAAAUUUAACUAAU ------------....((((.....(..(((((((((...---....)))))))))..)..-..)))).---.................((....))..--.............. ( -16.40, z-score = -2.14, R) >droSim1.chr2L 6823255 94 + 22036055 ------------CCUUUGCUAAAAAUUGGCAAUGAUCCCC---AAAUGAUCAUUGUUGACU-UAAGCAU---UUCUACUUUAUAUACUUGCAGAUGCCA--AAAAUUUAACUAAU ------------....((((.....(..(((((((((...---....)))))))))..)..-..)))).---.................((....))..--.............. ( -16.40, z-score = -2.14, R) >triCas2.chrUn_1 6536 100 - 1280983 ------------GCGUUUUGAUAAAUAUGCGACGAUACGCAAAGAAUUACCGCUUACAAUUACCAAUUUGCGUUCCAUCUUAUAUUUUUGUGAACGCUG--CGAAUACGGUUAA- ------------((((((....(((((((.((.(..(((((((.......................)))))))..).)).)))))))....))))))..--.............- ( -20.20, z-score = -0.77, R) >consensus ____________CCAUUGCUAUAAAUUGGCAAUGAUCCCC___AAAUGAUCAUUGUUGACU_UAAGCAU___UUCUAUUUUAUAUACUUGCAGAUACCA__AAAAUUUAACUAAU .................(((.....(..(((((((((..........)))))))))..).....)))................................................ (-13.40 = -13.30 + -0.10)
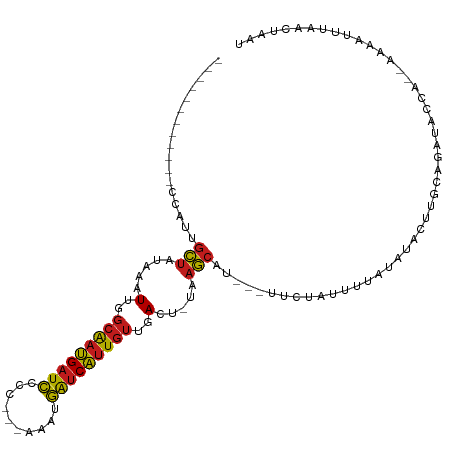
| Location | 7,035,000 – 7,035,094 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.85 |
| Shannon entropy | 0.46071 |
| G+C content | 0.30585 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -10.56 |
| Energy contribution | -10.03 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7035000 94 - 23011544 AUUAGUUAAAUUUU--UGGCAUCUGCAAGUAUAUAAAGUAGAA---AUGCUUA-AGUCAACAAUGAUCAUUU---GGGGAUCAUUGCCAAUUUUUAGCAAAGG------------ ....((((((...(--((((((((.(((((.....(((((...---.))))).-.((((....)))).))))---).)))....))))))..)))))).....------------ ( -24.40, z-score = -3.26, R) >droGri2.scaffold_15126 610435 104 - 8399593 AUUAGUUAAAUUUUU-UGGUAUCUGCAAGUAUAUAAAAUAGAA---AUGCUUA-AGUCAACAAUGAUCGUCUU--ACAGAUCAUUGCCAAUUUAUAGCAAUAGAGCAGUAG---- ...............-......((((...(((.....)))...---.((((((-(((...((((((((.....--...))))))))...))))).)))).....))))...---- ( -18.90, z-score = -1.11, R) >droMoj3.scaffold_6500 16207925 107 + 32352404 AUUAGUUAAA-UUUU-UGGUAUCUGCAAGUAUAUAAAAUAGAA---AUGCUUA-AGUCAACAAUGAUCGGUUU--ACAGAUCAUUGCCAAUUUACAGCAAUAUAGCAAUAGAGUA ....((((.(-((((-((.(((.((....)).)))...)))))---))(((((-(((...((((((((.....--...))))))))...))))).)))....))))......... ( -18.80, z-score = -0.90, R) >droVir3.scaffold_12723 2924079 104 + 5802038 AUUAGUUAAAUUUUU-UGGUAUCUGCAAGUACAUAAAAUAGAA---AUGCUUA-AGUCAACAAUGAUCGCUUC--ACAGAUCAUUGCCAAUUUAGAGCAAUAGAGCAGUAG---- ....(((..(((((.-..((((......))))..)))))....---.((((((-(((...((((((((.....--...))))))))...)))).)))))....))).....---- ( -19.40, z-score = -1.03, R) >droWil1.scaffold_180708 5106609 99 - 12563649 AUUAGUUAAAUUUUUGUGGUAUCUGCAAGUAUAUAAAAUAGAA---AUGCUUA-AGUCAACAAUGAUCAUUGUUGAGGGAUCAUUGCCAAUUUAGAGCAAAGU------------ .........((((((((..(((.((....)).)))..))))))---))((((.-..((((((((....))))))))((.(....).))......)))).....------------ ( -18.70, z-score = -0.84, R) >droPer1.super_5 1603111 98 - 6813705 AUUAGUUAAAUUUUU-GUGUAUCUGCAAGUAUAUAAAAUAGAA---AUGCUUA-AGUCAACAAUGAUCAUGG---G-GGAUCAUUGCCAAUUUAUAGCCAUAAUAGA-------- ....((((...((((-((((((......)))))))))).....---..(((((-(((...((((((((....---.-.))))))))...))))).)))..))))...-------- ( -18.40, z-score = -0.65, R) >dp4.chr4_group1 1668128 98 + 5278887 AUUAGUUAAAUUUUU-GUGUAUCUGCAAGUAUAUAAAAUAGAA---AUGCUUA-AGUCAACAAUGAUCAUGG---G-GGAUCAUUGCCAAUUUAUAGCCAUAAUAGA-------- ....((((...((((-((((((......)))))))))).....---..(((((-(((...((((((((....---.-.))))))))...))))).)))..))))...-------- ( -18.40, z-score = -0.65, R) >droAna3.scaffold_12916 106413 92 - 16180835 AUUAGUUAAAUUUU---GGAAUCUGCAAGUAUAUAAAAUAGAA---AUGCUUA-AGUCAACAAUGACCAUGGC--G-GGAUCAUUGCCAAUUUAUAGCAAGU------------- ....((((.(..((---((...((..((((((...........---)))))).-))....((((((((.....--.-)).))))))))))..).))))....------------- ( -16.40, z-score = -0.45, R) >droEre2.scaffold_4929 15945551 99 - 26641161 AUUAGUUAAAUUUU--UGGCAUCUGCAAGUAUAUAAAGUAGAA---AUGCUUA-AGUCAACAAUGAUCAUUU---GGGGAUCAUUGCCAAUUUUUAGCAGUUUUUAGG------- ....((((((...(--((((((((.(((((.....(((((...---.))))).-.((((....)))).))))---).)))....))))))..))))))..........------- ( -24.40, z-score = -2.97, R) >droYak2.chr2L 16445868 93 + 22324452 AUUAGUUAAAUUUU--UGGCAUCUGCAAGUAUAUAAAGUAGAA---AUGCUUA-AGUCAACAAUGAUCAUUU---GGGGAUCAUUGCCAAUUUUUAGCAAGG------------- ....((((((...(--((((((((.(((((.....(((((...---.))))).-.((((....)))).))))---).)))....))))))..))))))....------------- ( -24.40, z-score = -3.30, R) >droSec1.super_3 2559980 94 - 7220098 AUUAGUUAAAUUUU--UGGCAUCUGCAAGUAUAUAAAGUAGAA---AUGCUUA-AGUCAACAAUGAUCAUUU---GGGGAUCAUUGCCAAUUUUUAGCAAAGG------------ ....((((((...(--((((((((.(((((.....(((((...---.))))).-.((((....)))).))))---).)))....))))))..)))))).....------------ ( -24.40, z-score = -3.26, R) >droSim1.chr2L 6823255 94 - 22036055 AUUAGUUAAAUUUU--UGGCAUCUGCAAGUAUAUAAAGUAGAA---AUGCUUA-AGUCAACAAUGAUCAUUU---GGGGAUCAUUGCCAAUUUUUAGCAAAGG------------ ....((((((...(--((((((((.(((((.....(((((...---.))))).-.((((....)))).))))---).)))....))))))..)))))).....------------ ( -24.40, z-score = -3.26, R) >triCas2.chrUn_1 6536 100 + 1280983 -UUAACCGUAUUCG--CAGCGUUCACAAAAAUAUAAGAUGGAACGCAAAUUGGUAAUUGUAAGCGGUAAUUCUUUGCGUAUCGUCGCAUAUUUAUCAAAACGC------------ -.............--..(((((.....((((((..(((((.(((((((..(((.((((....)))).))).))))))).)))))..)))))).....)))))------------ ( -22.80, z-score = -1.39, R) >consensus AUUAGUUAAAUUUU__UGGCAUCUGCAAGUAUAUAAAAUAGAA___AUGCUUA_AGUCAACAAUGAUCAUUU___GGGGAUCAUUGCCAAUUUAUAGCAAUGG____________ .....................((((.............))))......(((...(((...((((((((..........))))))))...)))...)))................. (-10.56 = -10.03 + -0.54)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:49 2011