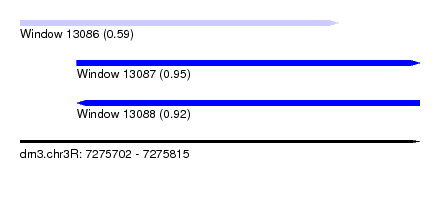
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,275,702 – 7,275,815 |
| Length | 113 |
| Max. P | 0.954009 |

| Location | 7,275,702 – 7,275,792 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 72.80 |
| Shannon entropy | 0.54867 |
| G+C content | 0.45123 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -11.23 |
| Energy contribution | -10.75 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.592516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
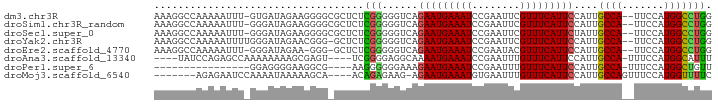
>dm3.chr3R 7275702 90 + 27905053 AAAGGCCAAAAAUUU-GUGAUAGAAGGGGCGCUCUCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCA--UUCCAUGGCCUGG ..((((((.......-(..((.((((..(((...(((((..((.....))..)))))...)))..).))).))..)..--.....)))))).. ( -30.24, z-score = -1.75, R) >droSim1.chr3R_random 441001 90 + 1307089 AAAGGCCAAAAAUUU-GGGAUAGAAGGGGCGCUCUCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCA--UUCCAUGGCCUGG ..((((((......(-(((((.((((..(((...(((((..((.....))..)))))...)))..).))).))).)))--.....)))))).. ( -30.50, z-score = -1.41, R) >droSec1.super_0 6432764 90 - 21120651 AAAGGCCAAAAAUUU-GGGAUAGAAGGGGCGCUCUCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCUAUUGCCA--UUCCAUGGCCUGG ..((((((......(-((((((((((..(((...(((((..((.....))..)))))...)))..).))))))).)))--.....)))))).. ( -35.00, z-score = -3.17, R) >droYak2.chr3R 11309498 90 + 28832112 AAAGGCCAAAAAUUUUGGGAUAGAACGGG-GCUCUCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCA--UUCCAUGGCCUGG ..((((((.......((((((..((((((-....(((((..((.....))..))))).))))))..))))))......--.....)))))).. ( -29.97, z-score = -1.48, R) >droEre2.scaffold_4770 3401835 88 - 17746568 AAAGGCCAAAAAUUU-GGGAUAGAA-GGG-GCUCUCGGGGGUCAGAAUGAAAUCCGAAUACGUUUCAUUCCAUUGCCA--UUCCAUGGCCUGG ..((((((......(-(((((....-(((-((((....))))).(((((((((........))))))))).....)))--))))))))))).. ( -27.80, z-score = -1.08, R) >droAna3.scaffold_13340 13710078 84 + 23697760 ----UAUCCAGAGCCAAAAAAAAGCGAGU----UCGGGGAGGCAAAAUGAAAUCCGAAUUUGUUUCAUUCCAUUGCCA-UUUCCAUGGCAUUU ----........((((.........(...----.)((..((((((((((((((........))))))))...))))).-)..)).)))).... ( -21.90, z-score = -1.59, R) >droPer1.super_6 900636 72 + 6141320 ----------------GGAGGGGAAGGCG----AAGGGGGGAAAGAAUGAAAUCCGAAUUUGUUUCAUUCCAUUGCCA-UUUCCAUGGCUGUU ----------------...((..(.((((----(.((((.((((.((............)).)))).)))).))))).-)..))......... ( -20.00, z-score = -0.29, R) >droMoj3.scaffold_6540 24755786 81 + 34148556 -------AGAGAAUCCAAAAUAAAAAGCA----ACAGAGAAG-AGAAUGAAAUGUGAAUUUGUUUCAUUCCAUUGCCAGUUUCCAUGGUUUUC -------.(((((.(((....((...(((----((......)-.((((((((((......))))))))))..))))...))....)))))))) ( -13.80, z-score = -0.81, R) >consensus AAAGGCCAAAAAUUU_GGGAUAGAAGGGG_GCUCUCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCA__UUCCAUGGCCUGG ...................................(((......(((((((((........)))))))))....((((.......))))))). (-11.23 = -10.75 + -0.48)
| Location | 7,275,718 – 7,275,815 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 62.84 |
| Shannon entropy | 0.73194 |
| G+C content | 0.47030 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -11.21 |
| Energy contribution | -10.96 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.954009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
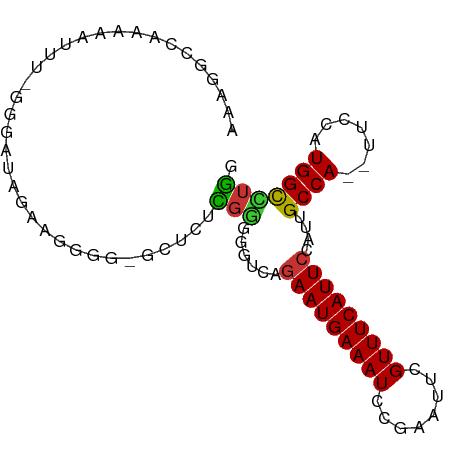
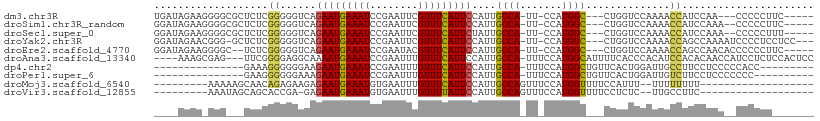
>dm3.chr3R 7275718 97 + 27905053 UGAUAGAAGGGGCGCUCUCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCA-UU-CCAUGGC---CUGGUCCAAAACCAUCCAA---CCCCCUUC----- .....(((((((.......((((..(((((((((((........)))))))))....((((-(.-..)))))---.))..))....))......---.)))))))----- ( -29.74, z-score = -0.86, R) >droSim1.chr3R_random 441017 98 + 1307089 GGAUAGAAGGGGCGCUCUCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCA-UU-CCAUGGC---CUGGUCCAAAACCAUCCAAA--CCCCCUUC----- .....(((((((((....)).((..(((((((((((........)))))))))....((((-(.-..)))))---.))..)).............--.)))))))----- ( -29.50, z-score = -0.48, R) >droSec1.super_0 6432780 98 - 21120651 GGAUAGAAGGGGCGCUCUCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCUAUUGCCA-UU-CCAUGGC---CUGGUCCAAAACCAUCCAAA--CCCCCUUU----- .....(((((((.......((.(((.((((((((((........))))))))))))).)).-..-...(((.---.((((.....)))).)))..--.)))))))----- ( -29.70, z-score = -0.70, R) >droYak2.chr3R 11309515 101 + 28832112 GGAUAGAACGGG-GCUCUCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCA-UU-CCAUGGC---CUGGUCCAAAACCAGCCAAAAUCCCCUCCUCC--- (((......(((-(.....((.(((..(((((((((........)))))))))....))).-..-)).((((---.((((.....))))))))....)))))))...--- ( -30.10, z-score = -0.54, R) >droEre2.scaffold_4770 3401851 98 - 17746568 GGAUAGAAGGGGC--UCUCGGGGGUCAGAAUGAAAUCCGAAUACGUUUCAUUCCAUUGCCA-UU-CCAUGGC---CUGGUCCAAAACCAGCCAACACCCCCCUUC----- .....(((((((.--....((.(((..(((((((((........)))))))))....))).-..-)).((((---.((((.....)))))))).....)))))))----- ( -33.90, z-score = -1.87, R) >droAna3.scaffold_13340 13710094 102 + 23697760 ----AAAGCGAG---UUCGGGGAGGCAAAAUGAAAUCCGAAUUUGUUUCAUUCCAUUGCCA-UUUCCAUGGCAUUUUCACCCACAUCCACACAACCAUCCUCUCCACUCC ----.....(((---(..(((((((...((((((((........))))))))....(((((-(....)))))).........................))))))))))). ( -25.90, z-score = -2.80, R) >dp4.chr2 25572907 85 + 30794189 ---------------GAAAGGGGGGAAGAAUGAAAUCCGAAUUUGUUUCAUUCCAUUGCCA-UUUCCAUGGCUGUUCACUGGAUUGCCUUCCUCCCCCACC--------- ---------------.....((((((.(((.(.((((((((.....)))........((((-(....)))))........))))).).))).))))))...--------- ( -28.50, z-score = -2.02, R) >droPer1.super_6 900648 84 + 6141320 ---------------GAAGGGGGGAAAGAAUGAAAUCCGAAUUUGUUUCAUUCCAUUGCCA-UUUCCAUGGCUGUUCACUGGAUUGUCUUCCUCCCCCCC---------- ---------------...(((((((..(((((((((........)))))))))....((((-(....)))))........(((......)))))))))).---------- ( -31.00, z-score = -2.86, R) >droMoj3.scaffold_6540 24755800 80 + 34148556 ---------AAAAAGCAACAGAGAAGAGAAUGAAAUGUGAAUUUGUUUCAUUCCAUUGCCAGUUUCCAUGGUUUUCCAUUU--UUUUUUUU------------------- ---------..........(((((((((((((.((((.((((.......))))))))((((.......))))....)))))--))))))))------------------- ( -14.80, z-score = -0.88, R) >droVir3.scaffold_12855 9727855 79 + 10161210 ---------AAAUAGCAGCACCGA-GAGAAUGAAAUGUGAAUUUGUUUUAUUCCAUUGCCAGUUUCCAUGGUUUUCCUCUC--UUGCCUUC------------------- ---------........(((..((-(((.....((((.((((.......))))))))((((.......))))....)))))--.)))....------------------- ( -15.60, z-score = -1.23, R) >consensus ____AGAA_GGG___UCUCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCA_UU_CCAUGGC___CUGGUCCAAAACCAUCCAAC__CCCCCU_______ ...................((......(((((((((........)))))))))....((((.......))))..............))...................... (-11.21 = -10.96 + -0.25)
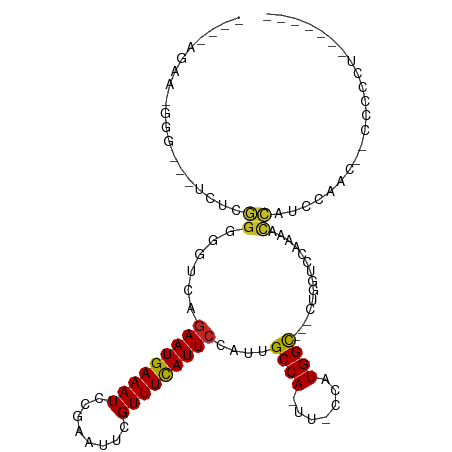
| Location | 7,275,718 – 7,275,815 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 62.84 |
| Shannon entropy | 0.73194 |
| G+C content | 0.47030 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -9.30 |
| Energy contribution | -9.42 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
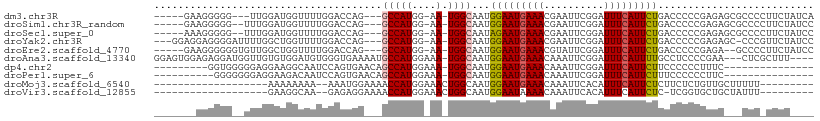
>dm3.chr3R 7275718 97 - 27905053 -----GAAGGGGG---UUGGAUGGUUUUGGACCAG---GCCAUGG-AA-UGGCAAUGGAAUGAAACGAAUUCGGAUUUCAUUCUGACCCCCGAGAGCGCCCCUUCUAUCA -----((((((((---((((..((((.........---(((((..-.)-))))...((((((((.((....))...)))))))))))).)))...)).)))))))..... ( -33.10, z-score = -0.72, R) >droSim1.chr3R_random 441017 98 - 1307089 -----GAAGGGGG--UUUGGAUGGUUUUGGACCAG---GCCAUGG-AA-UGGCAAUGGAAUGAAACGAAUUCGGAUUUCAUUCUGACCCCCGAGAGCGCCCCUUCUAUCC -----((((((((--(((((..((((.........---(((((..-.)-))))...((((((((.((....))...)))))))))))).))))..)).)))))))..... ( -34.80, z-score = -1.14, R) >droSec1.super_0 6432780 98 + 21120651 -----AAAGGGGG--UUUGGAUGGUUUUGGACCAG---GCCAUGG-AA-UGGCAAUAGAAUGAAACGAAUUCGGAUUUCAUUCUGACCCCCGAGAGCGCCCCUUCUAUCC -----.(((((((--(((((.(((((...))))).---(((((..-.)-))))..(((((((((.((....))...)))))))))....))))..)).))))))...... ( -32.20, z-score = -0.91, R) >droYak2.chr3R 11309515 101 - 28832112 ---GGAGGAGGGGAUUUUGGCUGGUUUUGGACCAG---GCCAUGG-AA-UGGCAAUGGAAUGAAACGAAUUCGGAUUUCAUUCUGACCCCCGAGAGC-CCCGUUCUAUCC ---..((((((((.((((((((((((...))))))---(((((..-.)-))))...((((((((.((....))...)))))))).....)))))).)-))).)))).... ( -36.80, z-score = -1.26, R) >droEre2.scaffold_4770 3401851 98 + 17746568 -----GAAGGGGGGUGUUGGCUGGUUUUGGACCAG---GCCAUGG-AA-UGGCAAUGGAAUGAAACGUAUUCGGAUUUCAUUCUGACCCCCGAGA--GCCCCUUCUAUCC -----((((((.(.(.((((((((((...))))))---(((((..-.)-))))...((((((((.((....))...)))))))).....)))).)--.)))))))..... ( -35.80, z-score = -1.10, R) >droAna3.scaffold_13340 13710094 102 - 23697760 GGAGUGGAGAGGAUGGUUGUGUGGAUGUGGGUGAAAAUGCCAUGGAAA-UGGCAAUGGAAUGAAACAAAUUCGGAUUUCAUUUUGCCUCCCCGAA---CUCGCUUU---- .((((...(.(((.(((...((((((.(((((.....((((((....)-))))).((........)).))))).).)))))...)))))).)..)---))).....---- ( -29.20, z-score = -0.92, R) >dp4.chr2 25572907 85 - 30794189 ---------GGUGGGGGAGGAAGGCAAUCCAGUGAACAGCCAUGGAAA-UGGCAAUGGAAUGAAACAAAUUCGGAUUUCAUUCUUCCCCCCUUUC--------------- ---------((.((((((((((.(.(((((........(((((....)-))))....((((.......))))))))).).))))))))))))...--------------- ( -36.40, z-score = -3.68, R) >droPer1.super_6 900648 84 - 6141320 ----------GGGGGGGAGGAAGACAAUCCAGUGAACAGCCAUGGAAA-UGGCAAUGGAAUGAAACAAAUUCGGAUUUCAUUCUUUCCCCCCUUC--------------- ----------((((((((((((((.(((((........(((((....)-))))....((((.......))))))))))).))))))))))))...--------------- ( -40.10, z-score = -5.12, R) >droMoj3.scaffold_6540 24755800 80 - 34148556 -------------------AAAAAAAA--AAAUGGAAAACCAUGGAAACUGGCAAUGGAAUGAAACAAAUUCACAUUUCAUUCUCUUCUCUGUUGCUUUUU--------- -------------------..(((((.--.((((((...(((.(....))))....(((((((((..........)))))))))....))))))..)))))--------- ( -14.70, z-score = -1.87, R) >droVir3.scaffold_12855 9727855 79 - 10161210 -------------------GAAGGCAA--GAGAGGAAAACCAUGGAAACUGGCAAUGGAAUAAAACAAAUUCACAUUUCAUUCUC-UCGGUGCUGCUAUUU--------- -------------------...((((.--(((((((...(((.(....)))).((((((((.......)))).))))...)))))-))..)))).......--------- ( -21.60, z-score = -3.05, R) >consensus _______AGGGGG__GUUGGAUGGUUUUGGACCAG___GCCAUGG_AA_UGGCAAUGGAAUGAAACAAAUUCGGAUUUCAUUCUGACCCCCGAGA___CCC_UUCU____ ......................................((((.......))))...(((((((((..........))))))))).......................... ( -9.30 = -9.42 + 0.12)
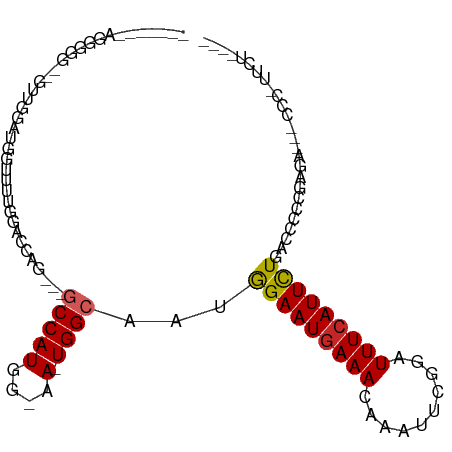
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:23 2011