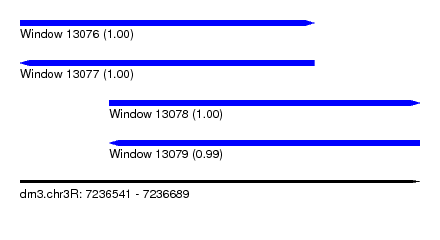
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,236,541 – 7,236,689 |
| Length | 148 |
| Max. P | 0.999975 |

| Location | 7,236,541 – 7,236,650 |
|---|---|
| Length | 109 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 61.52 |
| Shannon entropy | 0.80150 |
| G+C content | 0.42520 |
| Mean single sequence MFE | -30.39 |
| Consensus MFE | -18.83 |
| Energy contribution | -17.77 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.82 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.50 |
| SVM RNA-class probability | 0.999975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
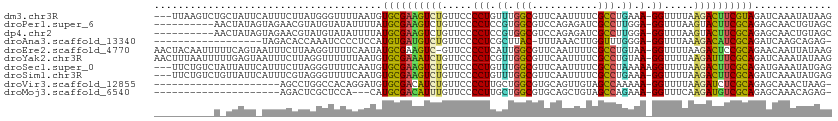
>dm3.chr3R 7236541 109 + 27905053 ---UUAAGUCUGCUAUUCAUUUCUUAUGGGUUUUAAUGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUGAAA-GGUUUUAAGACUUCGUAGAUCAAAUAUAAG ---....((((((.((((((.....))))))..........(((((((.....(((....((((.........))))....)-)).....))))))))))))).......... ( -28.20, z-score = -3.20, R) >droPer1.super_6 4506140 102 + 6141320 ----------AACUAUAGUAGAACGUAUGUAUAUUUUAUGCGAAGUCUGUUCCCCUCCGUGGCGUCCAGAGAUCGCCUUGGA-GGUUUAAGUACUUCGCAGAGCAACUGUAGC ----------..(((((((...((....))........((((((((((.....((((((.((((((....)).)))).))))-))....)).)))))))).....))))))). ( -36.10, z-score = -4.09, R) >dp4.chr2 29182159 102 + 30794189 ----------AACUAUAGUAGAACGUAUGUAUAUUUUAUGCGAAGUCUGUUCCCCUCCGUGGCGUCCAGAGAUCGCCUUGGA-GGUUUAAGUACUUCGCAGAGCAACUGUAGC ----------..(((((((...((....))........((((((((((.....((((((.((((((....)).)))).))))-))....)).)))))))).....))))))). ( -36.10, z-score = -4.09, R) >droAna3.scaffold_13340 13102338 92 - 23697760 ------------------UAGACACCAAAUCCCCUCCAUGUGAUGUCUGUUCCCCUCGCUUAC-UUUAAACUUGGUUUGGGA-GGUUUAAAGACAUCGCAGAUCAAGCAGAG- ------------------....................((((((((((.....((((.(..((-(........)))..).))-)).....))))))))))............- ( -25.50, z-score = -2.14, R) >droEre2.scaffold_4770 3360158 111 - 17746568 AACUACAAUUUUUCAGUAAUUUCUUAAGGUUUUCAAUAUGCGAAGUC-GUUCCCCUCAUUGGCGUUCAAUUUUCGCCUGUAA-GGUUUUAAGACUCCGCAGAACAAUUAUAAG ......................((((..((((((....((((.((((-.....(((.((.((((.........)))).)).)-))......)))).))))))).)))..)))) ( -21.70, z-score = -2.14, R) >droYak2.chr3R 11269378 112 + 28832112 AACUUUAAUUUUUGAGUAAUUUCUUAGGUUUUUUAAUGUGCGAAAUCUGUUCCCCUCGUUGGCGUUCAAUUUUCGCCUGUAA-GGUUUUAAGAUUUCGCAGAUCAAAUAUAAG .((((........)))).....((((.((((....((.((((((((((.....(((..(.((((.........)))).)..)-)).....)))))))))).)).)))).)))) ( -28.20, z-score = -3.69, R) >droSec1.super_0 6393241 110 - 21120651 ---UUCUGUCUAUUAUUCAUUUCUUAGGGUUUUCAAUGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUAAAAAGGUUUUAAGACUUCGCAGAUGAAAUAUGAG ---...................((((....(((((.(.((((((((((.....(((.(((((((.........)))).))).))).....)))))))))).))))))..)))) ( -31.80, z-score = -3.97, R) >droSim1.chr3R 13387354 109 - 27517382 ---UUCUGUCUGUUAUUCAUUUCGUAGGGUUUUCAAUGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUGAAA-GGUUUUAAGACUUCGCAGAUCAAAUAUGAG ---..................(((((.((((.......((((((((((.....(((....((((.........))))....)-)).....))))))))))))))...))))). ( -30.71, z-score = -3.27, R) >droVir3.scaffold_12855 6043451 90 + 10161210 ---------------------AGCCUGGCCACAGGAUGUGCGACAUCUGUUCCCCUUGCUGGCGUGCAGUUGUAGCCAAAAA-GGUUUUAAGAUCUCGCAGAGCAAACUAAG- ---------------------..((((....)))).(.(((((.((((.....((((..((((.(((....)))))))..))-)).....)))).))))).)..........- ( -30.20, z-score = -1.77, R) >droMoj3.scaffold_6540 6362645 87 + 34148556 ---------------------AGACUCGCUCCA---CAUGCGACAUUUGUUCCCCUUGCUGGCGUGCAGCUGUAGCCAGAAA-GGUUUCAAGAUGUCGCAGAGCAAACAGAG- ---------------------......((((..---..((((((((((.....((((.(((((.(((....)))))))).))-)).....))))))))))))))........- ( -35.40, z-score = -3.79, R) >consensus __________U_UUAUU_AUUACUUAAGGUUUUCAAUAUGCGAAGUCUGUUCCCCUCGUUGGCGUUCAAUUUUCGCCUGAAA_GGUUUUAAGACUUCGCAGAUCAAAUAUAAG ......................................((((((((((.....((..((.(((...........))).))...)).....))))))))))............. (-18.83 = -17.77 + -1.06)

| Location | 7,236,541 – 7,236,650 |
|---|---|
| Length | 109 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 61.52 |
| Shannon entropy | 0.80150 |
| G+C content | 0.42520 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -14.70 |
| Energy contribution | -14.37 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.08 |
| SVM RNA-class probability | 0.999944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7236541 109 - 27905053 CUUAUAUUUGAUCUACGAAGUCUUAAAACC-UUUCAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACAUUAAAACCCAUAAGAAAUGAAUAGCAGACUUAA--- (((((.((((((...((((((((.....((-(....((((.........))))....))).....))))))))...))))))....)))))...................--- ( -22.90, z-score = -2.84, R) >droPer1.super_6 4506140 102 - 6141320 GCUACAGUUGCUCUGCGAAGUACUUAAACC-UCCAAGGCGAUCUCUGGACGCCACGGAGGGGAACAGACUUCGCAUAAAAUAUACAUACGUUCUACUAUAGUU---------- ((((.(((.....((((((((.......((-(((..((((.((....))))))..))))).......))))))))........((....))...))).)))).---------- ( -31.14, z-score = -3.23, R) >dp4.chr2 29182159 102 - 30794189 GCUACAGUUGCUCUGCGAAGUACUUAAACC-UCCAAGGCGAUCUCUGGACGCCACGGAGGGGAACAGACUUCGCAUAAAAUAUACAUACGUUCUACUAUAGUU---------- ((((.(((.....((((((((.......((-(((..((((.((....))))))..))))).......))))))))........((....))...))).)))).---------- ( -31.14, z-score = -3.23, R) >droAna3.scaffold_13340 13102338 92 + 23697760 -CUCUGCUUGAUCUGCGAUGUCUUUAAACC-UCCCAAACCAAGUUUAAA-GUAAGCGAGGGGAACAGACAUCACAUGGAGGGGAUUUGGUGUCUA------------------ -((((((.......))(((((((.....((-((.........(((((..-.))))))))).....)))))))....)))).((((.....)))).------------------ ( -20.90, z-score = 0.68, R) >droEre2.scaffold_4770 3360158 111 + 17746568 CUUAUAAUUGUUCUGCGGAGUCUUAAAACC-UUACAGGCGAAAAUUGAACGCCAAUGAGGGGAAC-GACUUCGCAUAUUGAAAACCUUAAGAAAUUACUGAAAAAUUGUAGUU .((((((((.((((((((((((......((-(((..((((.........))))..))))).....-)))))))))............(((....)))..))).)))))))).. ( -30.80, z-score = -4.20, R) >droYak2.chr3R 11269378 112 - 28832112 CUUAUAUUUGAUCUGCGAAAUCUUAAAACC-UUACAGGCGAAAAUUGAACGCCAACGAGGGGAACAGAUUUCGCACAUUAAAAAACCUAAGAAAUUACUCAAAAAUUAAAGUU ......((((((.((((((((((.....((-((...((((.........))))...)))).....)))))))))).))))))............................... ( -27.50, z-score = -4.74, R) >droSec1.super_0 6393241 110 + 21120651 CUCAUAUUUCAUCUGCGAAGUCUUAAAACCUUUUUAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACAUUGAAAACCCUAAGAAAUGAAUAAUAGACAGAA--- ......(((((..((((((((((.....(((.(((.((((.........))))))).))).....))))))))))...)))))...........................--- ( -29.40, z-score = -4.38, R) >droSim1.chr3R 13387354 109 + 27517382 CUCAUAUUUGAUCUGCGAAGUCUUAAAACC-UUUCAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACAUUGAAAACCCUACGAAAUGAAUAACAGACAGAA--- ......((..((.((((((((((.....((-(....((((.........))))....))).....)))))))))).))..))............................--- ( -27.90, z-score = -3.85, R) >droVir3.scaffold_12855 6043451 90 - 10161210 -CUUAGUUUGCUCUGCGAGAUCUUAAAACC-UUUUUGGCUACAACUGCACGCCAGCAAGGGGAACAGAUGUCGCACAUCCUGUGGCCAGGCU--------------------- -............(((((.((((.....((-((.(((((...........))))).)))).....)))).)))))...((((....))))..--------------------- ( -26.50, z-score = -0.56, R) >droMoj3.scaffold_6540 6362645 87 - 34148556 -CUCUGUUUGCUCUGCGACAUCUUGAAACC-UUUCUGGCUACAGCUGCACGCCAGCAAGGGGAACAAAUGUCGCAUG---UGGAGCGAGUCU--------------------- -.....(((((((((((((((.(((...((-((((((((...........))))).)))))...))))))))))...---.))))))))...--------------------- ( -33.50, z-score = -2.62, R) >consensus CCUAUAUUUGAUCUGCGAAGUCUUAAAACC_UUUCAGGCGAAAAUUGAACGCCAACGAGGGGAACAGACUUCGCACAUUAAAAACCCUAAGAAAU_AAUAA_A__________ .............((((((((.......((.(((..(((...........)))..))).))......))))))))...................................... (-14.70 = -14.37 + -0.33)

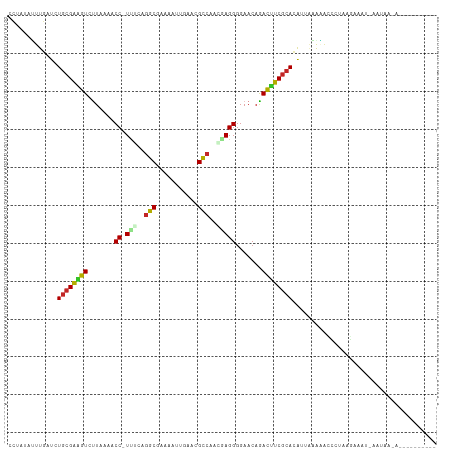
| Location | 7,236,574 – 7,236,689 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.49 |
| Shannon entropy | 0.55040 |
| G+C content | 0.45432 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -19.04 |
| Energy contribution | -18.02 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.997479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
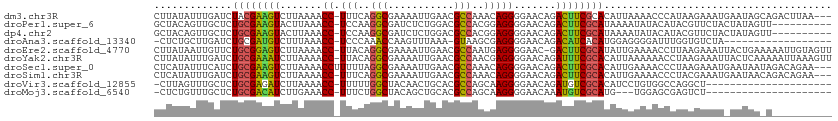
>dm3.chr3R 7236574 115 + 27905053 UGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUGAAA-GGUUUUAAGACUUCGUAGAUCAAAUAUA---AGCAAUCAUUCACCCGCAUCCAAGUGAAGCUCAAUU-GGUUC ..((((((((((.....(((....((((.........))))....)-)).....))))))))))((((((.....---(((.....(((((..........))))))))...))-)))). ( -30.30, z-score = -2.63, R) >droPer1.super_6 4506166 115 + 6141320 UAUGCGAAGUCUGUUCCCCUCCGUGGCGUCCAGAGAUCGCCUUGGA-GGUUUAAGUACUUCGCAGAGCAACUGUA---GCAAAUCAUUCACCUGCGUCAAAGUGAAGUUAAAUUUGGAU- ..((((((((((.....((((((.((((((....)).)))).))))-))....)).))))))))...........---.(((((..(((((.(......).))))).....)))))...- ( -36.00, z-score = -2.72, R) >dp4.chr2 29182185 115 + 30794189 UAUGCGAAGUCUGUUCCCCUCCGUGGCGUCCAGAGAUCGCCUUGGA-GGUUUAAGUACUUCGCAGAGCAACUGUA---GCAAAUCAUUCACCUGCGUCCAAGUGAAGUUAAAUUUGGAU- ..((((((((((.....((((((.((((((....)).)))).))))-))....)).))))))))..(((..((..---..........))..)))((((((((........))))))))- ( -40.70, z-score = -4.14, R) >droAna3.scaffold_13340 13102356 113 - 23697760 CAUGUGAUGUCUGUUCCCCUCGCUUAC-UUUAAACUUGGUUUGGGA-GGUUUAAAGACAUCGCAGAUCAAGCAGA---GCCA-UCAUUCACCCGCAUCCAAGUGAAGACAUAUU-GGAUC ..((((((((((.....((((.(..((-(........)))..).))-)).....))))))))))((((.....((---....-)).(((((..........)))))........-.)))) ( -31.20, z-score = -1.53, R) >droEre2.scaffold_4770 3360194 114 - 17746568 UAUGCGAAGUC-GUUCCCCUCAUUGGCGUUCAAUUUUCGCCUGUAA-GGUUUUAAGACUCCGCAGAACAAUUAUA---AGCAAUCAUUCACCCGCGUCCAAGUGAAGCUCAAUU-GGUUC ..((((.((((-.....(((.((.((((.........)))).)).)-))......)))).))))((((.......---(((.....(((((..........)))))))).....-.)))) ( -26.22, z-score = -1.70, R) >droYak2.chr3R 11269414 115 + 28832112 UGUGCGAAAUCUGUUCCCCUCGUUGGCGUUCAAUUUUCGCCUGUAA-GGUUUUAAGAUUUCGCAGAUCAAAUAUA---AGCAAUCAUUCACCCGCGUCCAAGUGAAGCACAAUU-GGUUC ..((((((((((.....(((..(.((((.........)))).)..)-)).....))))))))))..........(---(.((((..(((((..........))))).....)))-).)). ( -28.60, z-score = -2.16, R) >droSec1.super_0 6393274 116 - 21120651 UGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUAAAAAGGUUUUAAGACUUCGCAGAUGAAAUAUG---AGCAAUCAUUCACCCGCAUCCAAGUGAAGCUCAAUU-GGUUC ..((((((((((.....(((.(((((((.........)))).))).))).....)))))))))).........((---(((.....(((((..........))))))))))...-..... ( -34.60, z-score = -3.33, R) >droSim1.chr3R 13387387 115 - 27517382 UGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUGAAA-GGUUUUAAGACUUCGCAGAUCAAAUAUG---AGCAAUCAUUCACCCGCAUCCAAGUGAAGCUCAAUU-GGUUC ..((((((((((.....(((....((((.........))))....)-)).....))))))))))((((((...((---(((.....(((((..........)))))))))).))-)))). ( -37.40, z-score = -4.04, R) >droVir3.scaffold_12855 6043466 115 + 10161210 UGUGCGACAUCUGUUCCCCUUGCUGGCGUGCAGUUGUAGCCAAAAA-GGUUUUAAGAUCUCGCAGAGCAAACUAA---GCAAAUGUUUCACUCGCAACCAAUCGCAGUUGUAUUGGGCU- ..(((((.((((.....((((..((((.(((....)))))))..))-)).....)))).)))))(((.((((...---......))))..)))((..(((((.((....))))))))).- ( -32.50, z-score = -0.75, R) >droMoj3.scaffold_6540 6362657 115 + 34148556 CAUGCGACAUUUGUUCCCCUUGCUGGCGUGCAGCUGUAGCCAGAAA-GGUUUCAAGAUGUCGCAGAGCAAACAGA---GCAACUGUUUCACCCGCAUCCAACUGAAGUACUAUUGGGUU- ..((((((((((.....((((.(((((.(((....)))))))).))-)).....))))))))))..((((((((.---....)))))).....))((((((..(.....)..)))))).- ( -38.40, z-score = -2.10, R) >droGri2.scaffold_14906 6155445 114 - 14172833 UUUGUGGAAUCUGUUCCCCUGGUUGGCGUUGAGUUGUAGCCAAAAA-GGUUUUAAGAUUUCGCACAGUAAACUGG---GCAAAUGUU-CACUCGCAUCCAACUCGAGUUGUUGUCGAAU- ..((((((((((.....(((..(((((...........)))))..)-)).....))))))))))(((....)))(---((((.....-.(((((.........)))))..)))))....- ( -33.20, z-score = -1.84, R) >droWil1.scaffold_181089 4246511 118 + 12369635 UGUGCGAAACCUGUUCCCCUUGCUGGCGUGCAGUGAUAGCCUUCAA-GGUUUUAAGGUUUCGCAAAUCAAAUGUACAGAGCAAAAAUUCACCCGCCUCCAAGUGAAGCACUAUU-GGUCA ..((((((((((.....(((((..(((...........)))..)))-)).....)))))))))).............((.(((...(((((..........)))))......))-).)). ( -34.70, z-score = -2.15, R) >consensus UGUGCGAAGUCUGUUCCCCUCGUUGGCGUUCAAUUUUCGCCUGAAA_GGUUUUAAGACUUCGCAGAUCAAAUAUA___ACCAAUCAUUCACCCGCAUCCAAGUGAAGCUCAAUU_GGUUC ..((((((((((....((......(((...........)))......)).....))))))))))........................................................ (-19.04 = -18.02 + -1.02)

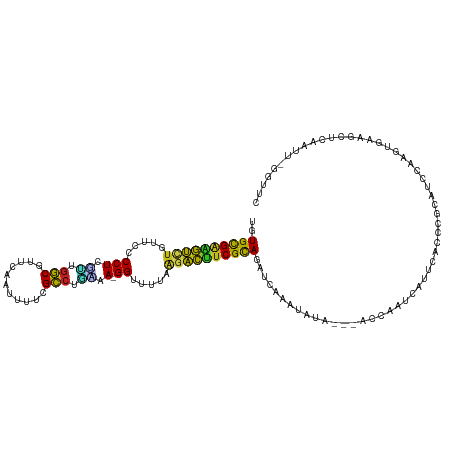
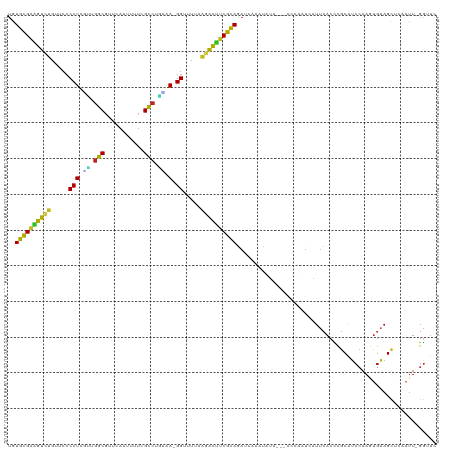
| Location | 7,236,574 – 7,236,689 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.49 |
| Shannon entropy | 0.55040 |
| G+C content | 0.45432 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -14.32 |
| Energy contribution | -13.68 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7236574 115 - 27905053 GAACC-AAUUGAGCUUCACUUGGAUGCGGGUGAAUGAUUGCU---UAUAUUUGAUCUACGAAGUCUUAAAACC-UUUCAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACA ....(-((.(((((((((((((....)))))))).....)))---))...))).....((((((((.....((-(....((((.........))))....))).....)))))))).... ( -31.90, z-score = -2.67, R) >droPer1.super_6 4506166 115 - 6141320 -AUCCAAAUUUAACUUCACUUUGACGCAGGUGAAUGAUUUGC---UACAGUUGCUCUGCGAAGUACUUAAACC-UCCAAGGCGAUCUCUGGACGCCACGGAGGGGAACAGACUUCGCAUA -...((((((....(((((((......))))))).)))))).---...........((((((((.......((-(((..((((.((....))))))..))))).......)))))))).. ( -37.94, z-score = -3.55, R) >dp4.chr2 29182185 115 - 30794189 -AUCCAAAUUUAACUUCACUUGGACGCAGGUGAAUGAUUUGC---UACAGUUGCUCUGCGAAGUACUUAAACC-UCCAAGGCGAUCUCUGGACGCCACGGAGGGGAACAGACUUCGCAUA -...((((((....((((((((....)))))))).)))))).---...........((((((((.......((-(((..((((.((....))))))..))))).......)))))))).. ( -41.14, z-score = -4.22, R) >droAna3.scaffold_13340 13102356 113 + 23697760 GAUCC-AAUAUGUCUUCACUUGGAUGCGGGUGAAUGA-UGGC---UCUGCUUGAUCUGCGAUGUCUUUAAACC-UCCCAAACCAAGUUUAAA-GUAAGCGAGGGGAACAGACAUCACAUG ((((.-.....(((((((((((....)))))))).))-)(((---...))).))))((.(((((((.....((-((.........(((((..-.))))))))).....))))))).)).. ( -27.80, z-score = -0.25, R) >droEre2.scaffold_4770 3360194 114 + 17746568 GAACC-AAUUGAGCUUCACUUGGACGCGGGUGAAUGAUUGCU---UAUAAUUGUUCUGCGGAGUCUUAAAACC-UUACAGGCGAAAAUUGAACGCCAAUGAGGGGAAC-GACUUCGCAUA ((((.-...(((((((((((((....)))))))).....)))---)).....))))(((((((((......((-(((..((((.........))))..))))).....-))))))))).. ( -39.80, z-score = -4.57, R) >droYak2.chr3R 11269414 115 - 28832112 GAACC-AAUUGUGCUUCACUUGGACGCGGGUGAAUGAUUGCU---UAUAUUUGAUCUGCGAAAUCUUAAAACC-UUACAGGCGAAAAUUGAACGCCAACGAGGGGAACAGAUUUCGCACA ....(-(((..(...(((((((....))))))))..))))..---...........((((((((((.....((-((...((((.........))))...)))).....)))))))))).. ( -33.10, z-score = -3.03, R) >droSec1.super_0 6393274 116 + 21120651 GAACC-AAUUGAGCUUCACUUGGAUGCGGGUGAAUGAUUGCU---CAUAUUUCAUCUGCGAAGUCUUAAAACCUUUUUAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACA (((..-...(((((((((((((....)))))))).....)))---))...)))...((((((((((.....(((.(((.((((.........))))))).))).....)))))))))).. ( -39.50, z-score = -4.52, R) >droSim1.chr3R 13387387 115 + 27517382 GAACC-AAUUGAGCUUCACUUGGAUGCGGGUGAAUGAUUGCU---CAUAUUUGAUCUGCGAAGUCUUAAAACC-UUUCAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACA ....(-((.(((((((((((((....)))))))).....)))---))...)))...((((((((((.....((-(....((((.........))))....))).....)))))))))).. ( -38.80, z-score = -4.06, R) >droVir3.scaffold_12855 6043466 115 - 10161210 -AGCCCAAUACAACUGCGAUUGGUUGCGAGUGAAACAUUUGC---UUAGUUUGCUCUGCGAGAUCUUAAAACC-UUUUUGGCUACAACUGCACGCCAGCAAGGGGAACAGAUGUCGCACA -((((((((.(......))))))..(((((((...)))))))---.......))).(((((.((((.....((-((.(((((...........))))).)))).....)))).))))).. ( -33.10, z-score = -1.04, R) >droMoj3.scaffold_6540 6362657 115 - 34148556 -AACCCAAUAGUACUUCAGUUGGAUGCGGGUGAAACAGUUGC---UCUGUUUGCUCUGCGACAUCUUGAAACC-UUUCUGGCUACAGCUGCACGCCAGCAAGGGGAACAAAUGUCGCAUG -...(((((.(.....).)))))....((((.((((((....---.))))))))))((((((((.(((...((-((((((((...........))))).)))))...))))))))))).. ( -39.10, z-score = -2.06, R) >droGri2.scaffold_14906 6155445 114 + 14172833 -AUUCGACAACAACUCGAGUUGGAUGCGAGUG-AACAUUUGC---CCAGUUUACUGUGCGAAAUCUUAAAACC-UUUUUGGCUACAACUCAACGCCAACCAGGGGAACAGAUUCCACAAA -..(((.((.((((....))))..)))))(((-....(((((---.(((....))).)))))((((.....((-(..(((((...........)))))..))).....))))..)))... ( -27.90, z-score = -1.54, R) >droWil1.scaffold_181089 4246511 118 - 12369635 UGACC-AAUAGUGCUUCACUUGGAGGCGGGUGAAUUUUUGCUCUGUACAUUUGAUUUGCGAAACCUUAAAACC-UUGAAGGCUAUCACUGCACGCCAGCAAGGGGAACAGGUUUCGCACA .....-.((((.((((((((((....)))))))).....)).))))..........((((((((((.....((-(((..(((...........)))..))))).....)))))))))).. ( -38.90, z-score = -2.55, R) >consensus GAACC_AAUUGAGCUUCACUUGGAUGCGGGUGAAUGAUUGCC___UAUAUUUGAUCUGCGAAGUCUUAAAACC_UUUCAGGCGAAAACUGAACGCCAACGAGGGGAACAGACUUCGCACA ..............((((.(((....))).))))......................((((((((.......((......(((...........))).....)).......)))))))).. (-14.32 = -13.68 + -0.64)
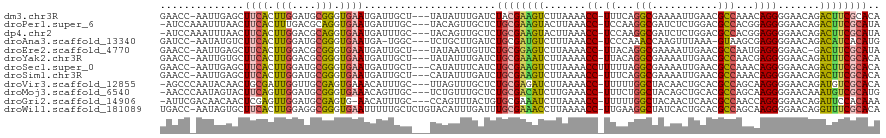
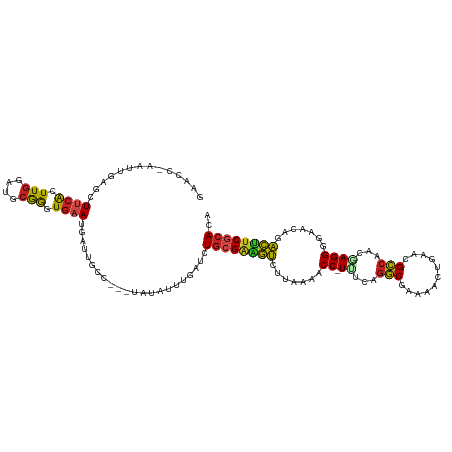
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:16 2011