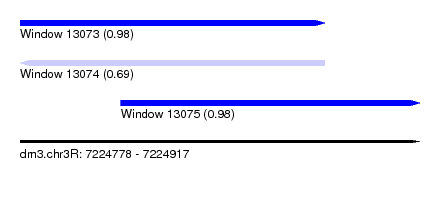
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,224,778 – 7,224,917 |
| Length | 139 |
| Max. P | 0.981512 |

| Location | 7,224,778 – 7,224,884 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.23 |
| Shannon entropy | 0.52350 |
| G+C content | 0.48696 |
| Mean single sequence MFE | -32.59 |
| Consensus MFE | -15.90 |
| Energy contribution | -14.61 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
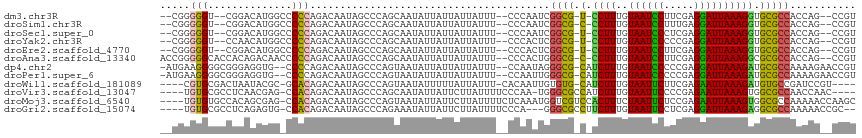
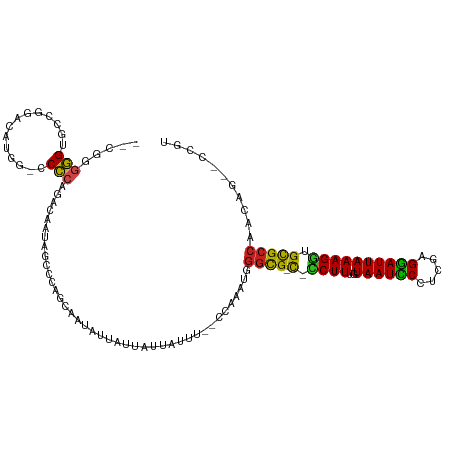
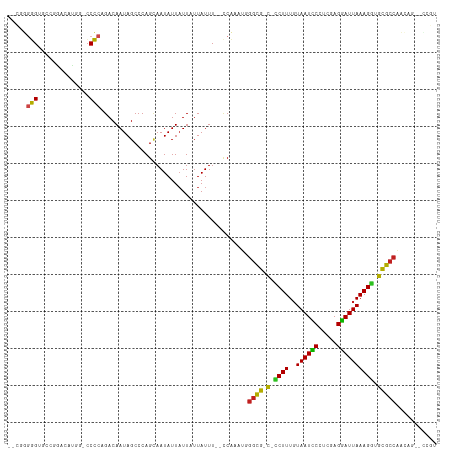
>dm3.chr3R 7224778 106 + 27905053 --CGGGGGU--CGGACAUGGCCCCCAGACAAUAGCCCAGCAAUAUUAUUAUUAUUU--CCCAAUCGGCG-U-CCUUUGUAAUCCUUCGAGGAUUAAAGGUGCGCCACCAG--CCGU --.((((((--((....)))))))).............((((((....))))....--.......((((-(-((((..((((((.....)))))))))).)))))....)--)... ( -35.80, z-score = -2.13, R) >droSim1.chr3R 13375627 106 - 27517382 --CGGGGGU--CGGACAUGGCCCCCAGACAAUAGCCCAGCAAUAUUAUUAUUAUUU--CCCAAUCGGCG-C-CCUUUGUAAUCCUUUGAGGAUUAAAGGUGCGCCACCAG--CCGU --.((((((--((....)))))))).............((((((....))))....--.......((((-(-((((..((((((.....)))))))))).)))))....)--)... ( -38.50, z-score = -2.68, R) >droSec1.super_0 6381516 106 - 21120651 --CGGGGGU--CGGACAUGGCCCCCAGACAAUAGCCCAGCAAUAUUAUUAUUAUUU--CCCAAUCGGCG-U-CCUUUGUAAUCCCUCGAGGAUUAAAGGUGCGCCACCAG--CCGU --.((((((--((....)))))))).............((((((....))))....--.......((((-(-((((..((((((.....)))))))))).)))))....)--)... ( -35.10, z-score = -1.79, R) >droYak2.chr3R 11257564 106 + 28832112 --CGGGGGU--CCAACAUGGCCCCCAGACAAUAGCCCAGCAAUAUUAUUAUUAUUU--CCCACUCGGCG-U-CCUUUGUAAUCCCCCGAGGAUUAAAGGUGCGCCACCAG--CCGU --.((((((--(......))))))).............((((((....))))....--.......((((-(-((((..((((((.....)))))))))).)))))....)--)... ( -33.80, z-score = -2.12, R) >droEre2.scaffold_4770 3348433 106 - 17746568 --CGGGGGU--CGGACAUGGCCCCCAGACAAUAGCCCAGCAAUAUUAUUAUUAUUU--CCCACUCGGCG-U-CCUUUGUAAUCCUUCGAGGAUUAAAGGUGCGCCACCAG--CCGU --.((((((--((....)))))))).............((((((....))))....--.......((((-(-((((..((((((.....)))))))))).)))))....)--)... ( -35.80, z-score = -2.06, R) >droAna3.scaffold_13340 13087669 110 - 23697760 ACCGGGGGCACCACAGACAACCCCCAGACAAUAGCCCAGCAAUAUUAUUAUUAUUU--CCCACUGGGCG-C-CCUUUGUAAUCCUUCGAGGAUUAAAGGCGCGCCACCAG--CCGU ...(((((............)))))........((((((.................--....))))))(-(-((((..((((((.....)))))))))).))........--.... ( -32.00, z-score = -1.70, R) >dp4.chr2 1596171 110 - 30794189 -AUGAAGGGGCGGGAGGUG--CCCCAGACAAUAGCCCAGUAAUAUUAUUAUUAUUU--CCAAUAGGGCG-CAUCUUUGUAAUCCCCCGAGGAUUAAAGAUGCGCCAAAAGAACCGU -.....((((((.....))--))))............(((((((....))))))).--.......((((-((((((..((((((.....))))))))))))))))........... ( -36.70, z-score = -2.75, R) >droPer1.super_6 1620688 110 - 6141320 -AUGAAGGGGCGGGAGGUG--CCCCAGACAAUAGCCCAGUAAUAUUAUUAUUAUUU--CCAAUUGGGCG-CAUCUUUGUAAUCCCCCGAGGAUUAAAGAUGCGCCAAAAGAACCGU -.....((((((.....))--))))...((((.(...(((((((....))))))).--.).))))((((-((((((..((((((.....))))))))))))))))........... ( -38.60, z-score = -3.09, R) >droWil1.scaffold_181089 8108152 105 - 12369635 ----CGUGCGACUAAUACGC-GCACAGACAAUAGCCCAGUAAUAUUUUUAUUAUUU-CACAAUUGUGUG-CAUCUUUGUAAUUCUUCGAGAAUUAAAGAUGUGCCGAUCCGU---- ----.(((((.(......))-))))..(((((.(...(((((((....))))))).-..).)))))(..-((((((..((((((.....))))))))))))..)........---- ( -26.10, z-score = -2.50, R) >droVir3.scaffold_13047 15991349 106 - 19223366 ----UGUGCGCCUCAACGAG-CCACAGACAAUAGCCCAGCAAUAUUAUUCUUAUUUUCCCAA-UGGGCGCCAUCUUUGUAAUUCCCCGAGAAUUAAAGUGGCGCCAACCAAC---- ----((((.((........)-)))))....................................-..(((((((.(((..((((((.....)))))))))))))))).......---- ( -24.20, z-score = -1.64, R) >droMoj3.scaffold_6540 13875625 111 - 34148556 ----UGUGUGCCACAGCGAG-CCACAGACAAUAGCCCAGUAAUAUUAUUCUUAUUUUCUCAAAUGGUCGUCCACUUUCUAAUUCUCCGAGAAUUAAAGUGGCGCCAAAAACCAAGC ----((.(((((((.((((.-(((.(((.(((((...((((....)))).))))).)))....)))))))........((((((.....))))))..))))))))).......... ( -24.10, z-score = -2.06, R) >droGri2.scaffold_15074 137575 106 - 7742996 ----UGUGCGCCUCAGAGUG-CCACAGACAAUAGCCCAGAAAUAUUAUUCUUAUUUUCCCA---GGGCGCCUUCUUUGUAAUUCCUCGAGGAUUAAAGAGGCGCCAAAAACCGC-- ----((.(((((((((.(((-((...((.((((....((((......)))))))).))...---.)))))))((((((........)))))).....)))))))))........-- ( -30.40, z-score = -2.46, R) >consensus __CGGGGGUGCCGGACAUGG_CCCCAGACAAUAGCCCAGCAAUAUUAUUAUUAUUU__CCAAAUGGGCG_C_CCUUUGUAAUCCCUCGAGGAUUAAAGGUGCGCCAACAG__CCGU .....(((..............)))........................................((((.(.((((..((((((.....)))))))))).)))))........... (-15.90 = -14.61 + -1.28)
| Location | 7,224,778 – 7,224,884 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.23 |
| Shannon entropy | 0.52350 |
| G+C content | 0.48696 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -17.74 |
| Energy contribution | -16.86 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7224778 106 - 27905053 ACGG--CUGGUGGCGCACCUUUAAUCCUCGAAGGAUUACAAAGG-A-CGCCGAUUGGG--AAAUAAUAAUAAUAUUGCUGGGCUAUUGUCUGGGGGCCAUGUCCG--ACCCCCG-- .(((--(...(((((..((((((((((.....))))))..))))-.-)))))((((..--...)))).........))))(((....))).(((((.(......)--.))))).-- ( -32.90, z-score = -0.30, R) >droSim1.chr3R 13375627 106 + 27517382 ACGG--CUGGUGGCGCACCUUUAAUCCUCAAAGGAUUACAAAGG-G-CGCCGAUUGGG--AAAUAAUAAUAAUAUUGCUGGGCUAUUGUCUGGGGGCCAUGUCCG--ACCCCCG-- .(((--(...((((((.((((((((((.....))))))..))))-)-)))))((((..--...)))).........))))(((....))).(((((.(......)--.))))).-- ( -37.90, z-score = -1.36, R) >droSec1.super_0 6381516 106 + 21120651 ACGG--CUGGUGGCGCACCUUUAAUCCUCGAGGGAUUACAAAGG-A-CGCCGAUUGGG--AAAUAAUAAUAAUAUUGCUGGGCUAUUGUCUGGGGGCCAUGUCCG--ACCCCCG-- .(((--(...(((((..((((((((((.....))))))..))))-.-)))))((((..--...)))).........))))(((....))).(((((.(......)--.))))).-- ( -33.00, z-score = 0.02, R) >droYak2.chr3R 11257564 106 - 28832112 ACGG--CUGGUGGCGCACCUUUAAUCCUCGGGGGAUUACAAAGG-A-CGCCGAGUGGG--AAAUAAUAAUAAUAUUGCUGGGCUAUUGUCUGGGGGCCAUGUUGG--ACCCCCG-- .(.(--((..(((((..((((((((((.....))))))..))))-.-)))))))).).--...................((((....))))(((((((.....))--.))))).-- ( -36.60, z-score = -0.60, R) >droEre2.scaffold_4770 3348433 106 + 17746568 ACGG--CUGGUGGCGCACCUUUAAUCCUCGAAGGAUUACAAAGG-A-CGCCGAGUGGG--AAAUAAUAAUAAUAUUGCUGGGCUAUUGUCUGGGGGCCAUGUCCG--ACCCCCG-- .(.(--((..(((((..((((((((((.....))))))..))))-.-)))))))).).--...................((((....))))(((((.(......)--.))))).-- ( -33.10, z-score = -0.14, R) >droAna3.scaffold_13340 13087669 110 + 23697760 ACGG--CUGGUGGCGCGCCUUUAAUCCUCGAAGGAUUACAAAGG-G-CGCCCAGUGGG--AAAUAAUAAUAAUAUUGCUGGGCUAUUGUCUGGGGGUUGUCUGUGGUGCCCCCGGU ..((--(.(((((((((((((((((((.....))))))...)))-)-)))((((..(.--..............)..)))))))))))))(((((((..........))))))).. ( -44.56, z-score = -1.97, R) >dp4.chr2 1596171 110 + 30794189 ACGGUUCUUUUGGCGCAUCUUUAAUCCUCGGGGGAUUACAAAGAUG-CGCCCUAUUGG--AAAUAAUAAUAAUAUUACUGGGCUAUUGUCUGGGG--CACCUCCCGCCCCUUCAU- .(((((((...((((((((((((((((.....))))))..))))))-)))).....))--)).(((((....))))))))(((....))).((((--(.......))))).....- ( -38.20, z-score = -2.76, R) >droPer1.super_6 1620688 110 + 6141320 ACGGUUCUUUUGGCGCAUCUUUAAUCCUCGGGGGAUUACAAAGAUG-CGCCCAAUUGG--AAAUAAUAAUAAUAUUACUGGGCUAUUGUCUGGGG--CACCUCCCGCCCCUUCAU- .(((((((...((((((((((((((((.....))))))..))))))-)))).....))--)).(((((....))))))))(((....))).((((--(.......))))).....- ( -38.20, z-score = -2.72, R) >droWil1.scaffold_181089 8108152 105 + 12369635 ----ACGGAUCGGCACAUCUUUAAUUCUCGAAGAAUUACAAAGAUG-CACACAAUUGUG-AAAUAAUAAAAAUAUUACUGGGCUAUUGUCUGUGC-GCGUAUUAGUCGCACG---- ----..((((.(((.((((((((((((.....))))))..))))))-((((....))))-.....................)))...))))((((-((......).))))).---- ( -23.80, z-score = -1.46, R) >droVir3.scaffold_13047 15991349 106 + 19223366 ----GUUGGUUGGCGCCACUUUAAUUCUCGGGGAAUUACAAAGAUGGCGCCCA-UUGGGAAAAUAAGAAUAAUAUUGCUGGGCUAUUGUCUGUGG-CUCGUUGAGGCGCACA---- ----.(..((.((((((((((((((((.....))))))..))).))))))).)-)..).................(((.(((((((.....))))-)))((....)))))..---- ( -32.50, z-score = -1.14, R) >droMoj3.scaffold_6540 13875625 111 + 34148556 GCUUGGUUUUUGGCGCCACUUUAAUUCUCGGAGAAUUAGAAAGUGGACGACCAUUUGAGAAAAUAAGAAUAAUAUUACUGGGCUAUUGUCUGUGG-CUCGCUGUGGCACACA---- .((..(....(((((((((((((((((.....))))))..)))))).)).))).)..))....................(((((((.....))))-)))((....)).....---- ( -29.50, z-score = -1.30, R) >droGri2.scaffold_15074 137575 106 + 7742996 --GCGGUUUUUGGCGCCUCUUUAAUCCUCGAGGAAUUACAAAGAAGGCGCCC---UGGGAAAAUAAGAAUAAUAUUUCUGGGCUAUUGUCUGUGG-CACUCUGAGGCGCACA---- --(((......(((((((((((..(((....))).....)))).)))))))(---(.(((.....((((......))))..(((((.....))))-)..))).)).)))...---- ( -33.10, z-score = -1.40, R) >consensus ACGG__CUGGUGGCGCACCUUUAAUCCUCGAAGGAUUACAAAGG_G_CGCCCAUUGGG__AAAUAAUAAUAAUAUUGCUGGGCUAUUGUCUGGGG_CCAUCUCCGGCACCCCCG__ ...........((((..((((((((((.....))))))..))))...))))............................((((....))))(((..............)))..... (-17.74 = -16.86 + -0.89)

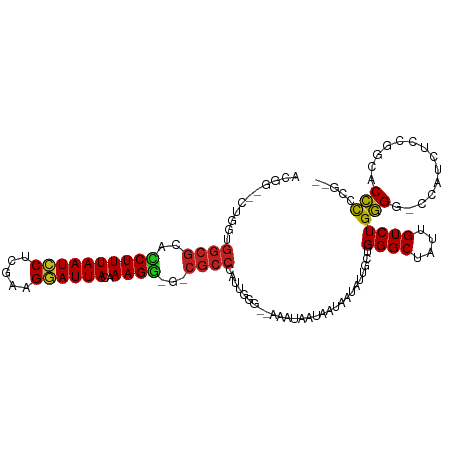
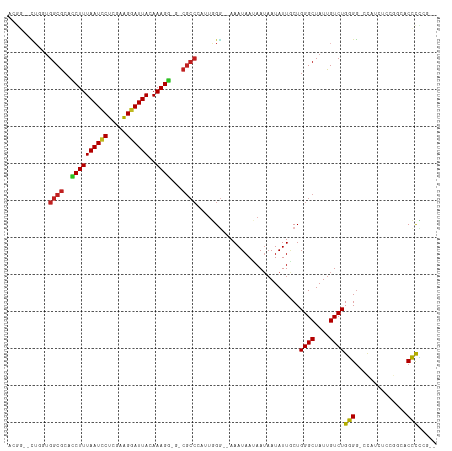
| Location | 7,224,813 – 7,224,917 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.24 |
| Shannon entropy | 0.42873 |
| G+C content | 0.40238 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -13.77 |
| Energy contribution | -13.13 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
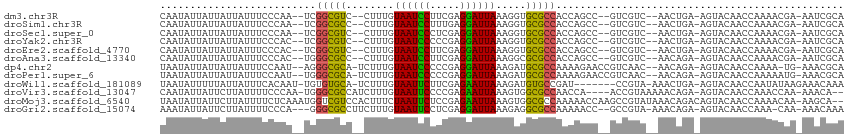
>dm3.chr3R 7224813 104 + 27905053 CAAUAUUAUUAUUAUUUCCCAA--UCGGCGUC--CUUUGUAAUCCUUCGAGGAUUAAAGGUGCGCCACCAGCC--GUCGUC--AACUGA-AGUACAACCAAAACGA-AAUCGCA ......................--..((((((--(((..((((((.....)))))))))).)))))....((.--.((((.--......-............))))-....)). ( -21.61, z-score = -1.42, R) >droSim1.chr3R 13375662 104 - 27517382 CAAUAUUAUUAUUAUUUCCCAA--UCGGCGCC--CUUUGUAAUCCUUUGAGGAUUAAAGGUGCGCCACCAGCC--GUCGUC--AACUGA-AGUACAACCAAAACGA-AAUCGCA ......................--..((((((--(((..((((((.....)))))))))).)))))....((.--.((((.--......-............))))-....)). ( -24.31, z-score = -2.19, R) >droSec1.super_0 6381551 104 - 21120651 CAAUAUUAUUAUUAUUUCCCAA--UCGGCGUC--CUUUGUAAUCCCUCGAGGAUUAAAGGUGCGCCACCAGCC--GUCGUC--AACUGA-AGUACAACCAAAACGA-AAUCGCA ......................--..((((((--(((..((((((.....)))))))))).)))))....((.--.((((.--......-............))))-....)). ( -20.91, z-score = -1.21, R) >droYak2.chr3R 11257599 104 + 28832112 CAAUAUUAUUAUUAUUUCCCAC--UCGGCGUC--CUUUGUAAUCCCCCGAGGAUUAAAGGUGCGCCACCAGCC--GUCGUC--AACUGA-AGUACAACCAAAACGA-AAUCGCA ......................--..((((((--(((..((((((.....)))))))))).)))))....((.--.((((.--......-............))))-....)). ( -20.91, z-score = -1.21, R) >droEre2.scaffold_4770 3348468 104 - 17746568 CAAUAUUAUUAUUAUUUCCCAC--UCGGCGUC--CUUUGUAAUCCUUCGAGGAUUAAAGGUGCGCCACCAGCC--GUCGUC--AACUGA-AGUACAACCAAAACGA-AAUCGCA ......................--..((((((--(((..((((((.....)))))))))).)))))....((.--.((((.--......-............))))-....)). ( -21.61, z-score = -1.36, R) >droAna3.scaffold_13340 13087708 104 - 23697760 CAAUAUUAUUAUUAUUUCCCAC--UGGGCGCC--CUUUGUAAUCCUUCGAGGAUUAAAGGCGCGCCACCAGCC--GUCGUC--AACAGA-AGUACAACCAAAACGA-AAUCGCA ......................--..((((((--(((..((((((.....)))))))))).)))))....((.--.((((.--......-............))))-....)). ( -24.01, z-score = -1.63, R) >dp4.chr2 1596207 106 - 30794189 UAAUAUUAUUAUUAUUUCCAAU--AGGGCGCA-UCUUUGUAAUCCCCCGAGGAUUAAAGAUGCGCCAAAAGAACCGUCAAC--AACAGA-AGUACAACCAAAA-UG-AAACGCA ............((((((....--..((((((-((((..((((((.....))))))))))))))))....((....))...--....))-)))).........-..-....... ( -25.60, z-score = -3.92, R) >droPer1.super_6 1620724 107 - 6141320 UAAUAUUAUUAUUAUUUCCAAU--UGGGCGCA-UCUUUGUAAUCCCCCGAGGAUUAAAGAUGCGCCAAAAGAACCGUCAAC--AACAGA-AGUACAACCAAAAAUG-AAACGCA ............((((((...(--((((((((-((((..((((((.....))))))))))))))))....((....))..)--))..))-))))............-....... ( -26.40, z-score = -4.00, R) >droWil1.scaffold_181089 8108186 103 - 12369635 UAAUAUUUUUAUUAUUUCACAAU-UGUGUGCA-UCUUUGUAAUUCUUCGAGAAUUAAAGAUGUGCCGAU-------CCGUA-AAACUGA-AGUACAACCAAUAUAAGAAACAAA ..(((((.....(((((((..((-((.(..((-((((..((((((.....))))))))))))..)))))-------..(..-...))))-)))).....))))).......... ( -18.60, z-score = -2.05, R) >droVir3.scaffold_13047 15991383 105 - 19223366 CAAUAUUAUUCUUAUUUUCCCAA-UGGGCGCCAUCUUUGUAAUUCCCCGAGAAUUAAAGUGGCGCCAACCA----ACCGUAAAAACAGA-AGUACAACCAAACCAA-AAACA-- ........((((...........-..(((((((.(((..((((((.....)))))))))))))))).....----...((....)))))-)...............-.....-- ( -19.60, z-score = -2.21, R) >droMoj3.scaffold_6540 13875659 111 - 34148556 UAAUAUUAUUCUUAUUUUCUCAAAUGGUCGUCCACUUUCUAAUUCUCCGAGAAUUAAAGUGGCGCCAAAAACCAAGCCGUAUAAACAGACAGUACAACCAAAACAA-AAGCA-- ........................(((.((.((((((..((((((.....)))))))))))))))))...........((((.........))))...........-.....-- ( -18.10, z-score = -2.36, R) >droGri2.scaffold_15074 137609 105 - 7742996 AAAUAUUAUUCUUAUUUUCCCA---GGGCGCCUUCUUUGUAAUUCCUCGAGGAUUAAAGAGGCGCCAAAAACC--GCCGUA-AAACAGA-AGUACAACCAAA-CAA-AAACAAA ......................---.(((((((.(((((....(((....))).)))))))))))).......--......-.......-............-...-....... ( -21.60, z-score = -2.07, R) >consensus CAAUAUUAUUAUUAUUUCCCAA__UGGGCGCC__CUUUGUAAUCCCUCGAGGAUUAAAGGUGCGCCAACAGCC__GUCGUC__AACAGA_AGUACAACCAAAACGA_AAUCGCA ..........................(((((...(((..((((((.....)))))))))..)))))................................................ (-13.77 = -13.13 + -0.64)

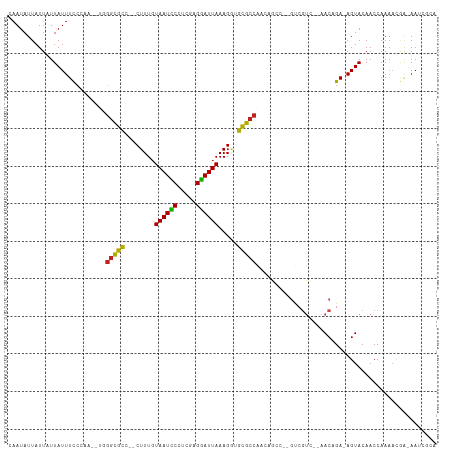
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:13 2011