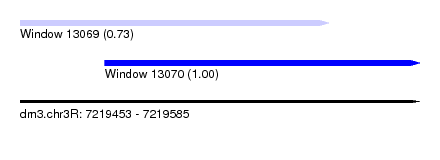
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,219,453 – 7,219,585 |
| Length | 132 |
| Max. P | 0.999925 |

| Location | 7,219,453 – 7,219,555 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 73.80 |
| Shannon entropy | 0.51836 |
| G+C content | 0.56148 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -20.24 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
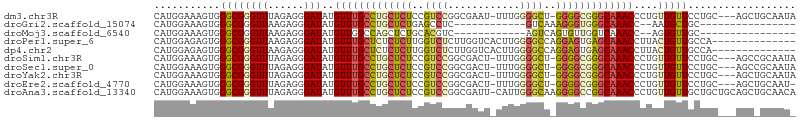
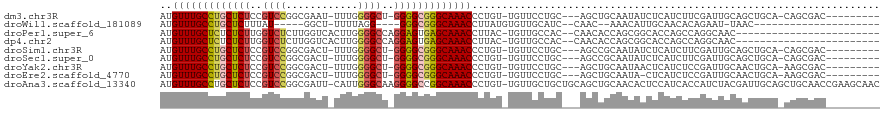
>dm3.chr3R 7219453 102 + 27905053 CAUGGAAAGUGGGCGGGUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGAAU-UUUGGGGCU-GGGGCGGGCAAACCCUGUUGUUCCUGC---AGCUGCAAUA .......(((..(((((..(((.(((....(((((((((((((..((((.(.....-..).)))).-)))))))))))))))).)))..)))))---.)))...... ( -40.80, z-score = -1.29, R) >droGri2.scaffold_15074 131981 77 - 7742996 CAUGGAAAGUGGGCGGGUUAAGAGGGAUAUGUUUGCCUGCUCUGAGCCUC------------GUCAAAGGGUGGGCAAACC--AAUGCUGC---------------- ..(((.....((((((((.((.(......).)).))))))))...(((.(------------..(....)..))))...))--).......---------------- ( -22.10, z-score = -0.03, R) >droMoj3.scaffold_6540 13869856 77 - 34148556 CAUGGAAAGUGGGCGGGUUAAGAGGGAUAUGUUGGCCAGCUCUGCACGUC------------AGUCAGUGUUGGUCAAACC--AGUGUUGC---------------- ..(((...(..(((.((((((.(......).)))))).)).)..)...))------------)..(((..((((.....))--))..))).---------------- ( -21.40, z-score = -0.51, R) >droPer1.super_6 1615209 93 - 6141320 CAUGGAGAGUGGGCGGGUUAAGAGGGAUAUGUUUGCUCUCUCUUGGUCUCUUGGUCACUUGGGGCCAGGAGUGAGCAAACCUUACUGUUGCCA-------------- ...........(((((....((..((....((((((((.(((((((((((..(....)..))))))))))).))))))))))..)).))))).-------------- ( -38.70, z-score = -2.81, R) >dp4.chr2 1590671 93 - 30794189 CAUGGAGAGUGGGCGGGUUAAGAGGGAUAUGUUUGCUCUCUCUUGGUCUCUUGGUCACUUGGGGCCAGGAGUGAGCAAACCUUACUGUUGCCA-------------- ...........(((((....((..((....((((((((.(((((((((((..(....)..))))))))))).))))))))))..)).))))).-------------- ( -38.70, z-score = -2.81, R) >droSim1.chr3R 13370341 102 - 27517382 CAUGGAAAGUGGGCGGGUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGACU-UUUGGGGCU-GGGGCGGGCAAACCCUGUUGUUCCUGC---AGCCGCAAUA ........(((((((((..(((.(((....(((((((((((((..((((.(.....-..).)))).-)))))))))))))))).)))..)))))---..)))).... ( -43.20, z-score = -1.48, R) >droSec1.super_0 6376249 102 - 21120651 CAUGGAAAGUGGGCGGGUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGACU-UUUGGGGCU-GGGGCGGGCAAACCCUGUUGUUCCUGC---AGCCGCAAUA ........(((((((((..(((.(((....(((((((((((((..((((.(.....-..).)))).-)))))))))))))))).)))..)))))---..)))).... ( -43.20, z-score = -1.48, R) >droYak2.chr3R 11252250 102 + 28832112 CAUGGAAAGUGGGCGGGUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGACU-UUUGGGGCU-GGGGCGGGCAAACCCUGUUGUUCCUGC---AGCUGCAAUA .......(((..(((((..(((.(((....(((((((((((((..((((.(.....-..).)))).-)))))))))))))))).)))..)))))---.)))...... ( -40.80, z-score = -1.08, R) >droEre2.scaffold_4770 3343176 101 - 17746568 CAUGGAAAGUGGGCGGGUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGACU-UUUGGGGCU-GGGGCGGGCAAACCCUGUUGUUCCUGC---AGCUGCAAU- .......(((..(((((..(((.(((....(((((((((((((..((((.(.....-..).)))).-)))))))))))))))).)))..)))))---.))).....- ( -40.80, z-score = -1.04, R) >droAna3.scaffold_13340 13082227 106 - 23697760 CAUGGAAAGUGGGCGGGUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGAUU-CAUUGGGCAAGGGGCCGGCAAACCCUGUUGUUGCUGCUGCAGCUGCAACA ...(((.((..((((((..((......))..))))))..)).)))((((((.....-..)))))).((((.........))))(((((.((((...)))).))))). ( -38.00, z-score = 0.37, R) >consensus CAUGGAAAGUGGGCGGGUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGACU_UUUGGGGCU_GGGGCGGGCAAACCCUGUUGUUCCUGC___AGC_GCAAU_ ...........((((((((......)))..(((((((((((((..((((............))))..)))))))))))))....))))).................. (-20.24 = -20.24 + 0.00)
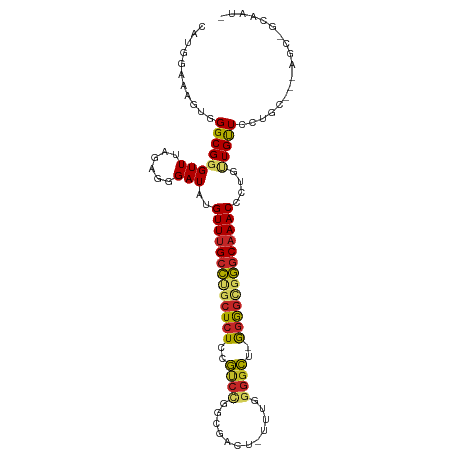
| Location | 7,219,481 – 7,219,585 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.18 |
| Shannon entropy | 0.62699 |
| G+C content | 0.55502 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.50 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.93 |
| SVM RNA-class probability | 0.999925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
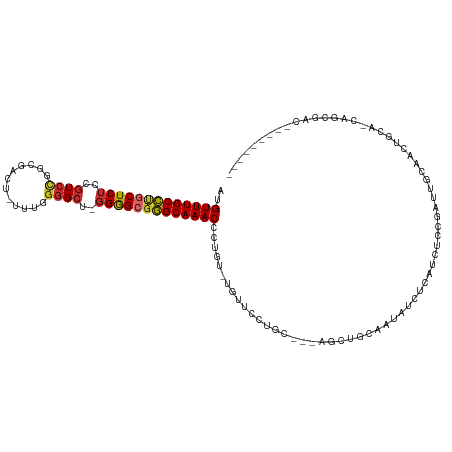
>dm3.chr3R 7219481 104 + 27905053 AUGUUUGCCUGCUCUCCGUCCGGCGAAU-UUUGGGGCU-GGGGCGGGCAAACCCUGU-UGUUCCUGC---AGCUGCAAUAUCUCAUCUUCGAUUGCAGCUGCA-CAGCGAC--------- ..(((((((((((((..((((.(.....-..).)))).-)))))))))))))...((-((((..(((---(((((((((............))))))))))))-.))))))--------- ( -48.50, z-score = -4.29, R) >droWil1.scaffold_181089 8102229 84 - 12369635 AUGUUUGCCUGCUCUUUAU-----GGCU-UUUUAGG----GGGCGGGCAAACCUUAUGUGUUGCAUC--CAAC--AAACAUUGCAACACAGAAU-UAAC--------------------- ..((((((((((((((((.-----....-...))))----))))))))))))....(((((((((..--....--......)))))))))....-....--------------------- ( -36.20, z-score = -5.92, R) >droPer1.super_6 1615237 93 - 6141320 AUGUUUGCUCUCUCUUGGUCUCUUGGUCACUUGGGGCCAGGAGUGAGCAAACCUUAC-UGUUGCCAC--CAACACCAGCGGCACCAGCCAGGCAAC------------------------ ..((((((((.(((((((((((..(....)..))))))))))).)))))))).....-.((((((..--...(....).(((....))).))))))------------------------ ( -43.10, z-score = -4.18, R) >dp4.chr2 1590699 93 - 30794189 AUGUUUGCUCUCUCUUGGUCUCUUGGUCACUUGGGGCCAGGAGUGAGCAAACCUUAC-UGUUGCCAC--CAACACCAGCGGCACCAGCCAGGCAAC------------------------ ..((((((((.(((((((((((..(....)..))))))))))).)))))))).....-.((((((..--...(....).(((....))).))))))------------------------ ( -43.10, z-score = -4.18, R) >droSim1.chr3R 13370369 104 - 27517382 AUGUUUGCCUGCUCUCCGUCCGGCGACU-UUUGGGGCU-GGGGCGGGCAAACCCUGU-UGUUCCUGC---AGCCGCAAUAUCUCAUCUUCGAUUGCAGCUGCA-CAGCGAC--------- ..(((((((((((((..((((.(.....-..).)))).-)))))))))))))...((-((((..(((---(((.(((((............))))).))))))-.))))))--------- ( -43.90, z-score = -2.74, R) >droSec1.super_0 6376277 104 - 21120651 AUGUUUGCCUGCUCUCCGUCCGGCGACU-UUUGGGGCU-GGGGCGGGCAAACCCUGU-UGUUCCUGC---AGCCGCAAUAUCUCAUCUUCGAUUGCAGCUGCA-CAGCGAC--------- ..(((((((((((((..((((.(.....-..).)))).-)))))))))))))...((-((((..(((---(((.(((((............))))).))))))-.))))))--------- ( -43.90, z-score = -2.74, R) >droYak2.chr3R 11252278 104 + 28832112 AUGUUUGCCUGCUCUCCGUCCGGCGACU-UUUGGGGCU-GGGGCGGGCAAACCCUGU-UGUUCCUGC---AGCUGCAAUAACUCAUCUCCGAUUGCAACUGCA-AAGCGAC--------- ..(((((((((((((..((((.(.....-..).)))).-)))))))))))))...((-((((..(((---((.((((((............)))))).)))))-.))))))--------- ( -42.70, z-score = -3.13, R) >droEre2.scaffold_4770 3343204 103 - 17746568 AUGUUUGCCUGCUCUCCGUCCGGCGACU-UUUGGGGCU-GGGGCGGGCAAACCCUGU-UGUUCCUGC---AGCUGCAAUA-CUCAUCUCCGAUUGCAACUGCA-AAGCGAC--------- ..(((((((((((((..((((.(.....-..).)))).-)))))))))))))...((-((((..(((---((.((((((.-(........))))))).)))))-.))))))--------- ( -43.20, z-score = -3.24, R) >droAna3.scaffold_13340 13082255 118 - 23697760 AUGUUUGCCUGCUCUCCGUCCGGCGAUU-CAUUGGGCAAGGGGCCGGCAAACCCUGU-UGUUGCUGCUGCAGCUGCAACACUCCAUCACCAUCUACGAUUGCAGCUGCAACCGAAGCAAC ..(((((((.(((((..((((((.....-..))))))..))))).))))))).....-.((((((..(((((((((((....................))))))))))).....)))))) ( -47.05, z-score = -2.90, R) >consensus AUGUUUGCCUGCUCUCCGUCCGGCGACU_UUUGGGGCU_GGGGCGGGCAAACCCUGU_UGUUCCUGC___AGCUGCAAUAUCUCAUCUCCGAUUGCAACUGCA_CAGCGAC_________ ..(((((((((((((..((((............))))..))))))))))))).................................................................... (-22.54 = -22.50 + -0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:09 2011