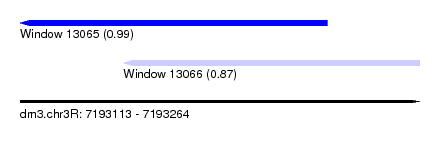
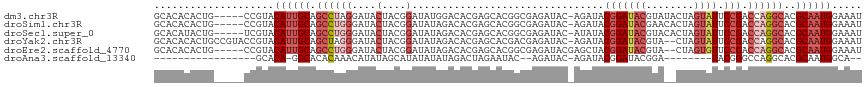
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,193,113 – 7,193,264 |
| Length | 151 |
| Max. P | 0.992486 |

| Location | 7,193,113 – 7,193,229 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.70 |
| Shannon entropy | 0.22016 |
| G+C content | 0.52156 |
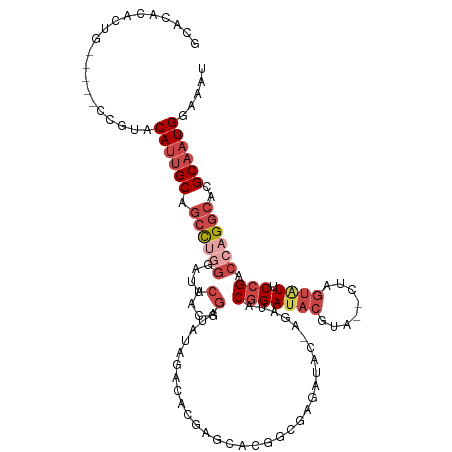
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -30.29 |
| Energy contribution | -31.93 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7193113 116 - 27905053 CGGAUAUGGACACGAGCACGGCGAGAUAC-AGAUACGGAUACGUAUACUAGUAUUCCGACCAGGCACGCAAUGGAAAUGGCCAGAAAAUGGCCAUAUGUGGCCAAGUCGGCUCCCAU .....((((....((((.(((((..((((-(((((((....))))).)).))))..).....(((.((((......(((((((.....))))))).)))))))..)))))))))))) ( -42.90, z-score = -3.80, R) >droSim1.chr3R 13356199 116 + 27517382 CGGAUAUAGACACGAGCACGGCGAGAUAC-AGAUACGGAUACGAACACUAGUAUUCCGACCAGGCACGCAAUGGAAAUGGCCAGAAAAUGGCCAUAUGUGGCCAAGUCGGCUCCCAC .............((((.((((.......-.....(((((((........))).))))....(((.((((......(((((((.....))))))).)))))))..)))))))).... ( -38.10, z-score = -3.45, R) >droSec1.super_0 6358776 116 + 21120651 CGGAUAUAGACACGAGCACGGCGAGAUAC-AUAUACGGAUACGUACACUAGUAUUCCGACCAGGCACGCAAUGGAAAUGGCCAGAAAAUGGCCAUAUGUGGCCAAGUCGGCUCCCGC (((..........((((.((((.......-.....(((((((........))).))))....(((.((((......(((((((.....))))))).)))))))..))))))))))). ( -38.20, z-score = -2.56, R) >droYak2.chr3R 11235072 114 - 28832112 CGGAUAUAGACACGAGCACGACGAGAUAC-AGAUACGGAUACG--UACUAGUAUUCCGACCAGGCACGCAAUGGAAAUGGCCAGAAAAUGGCCAUAUGUGGCCAAGUCGGCUCCCAC .............((((.(((((..((((-((.((((....))--)))).))))..).....(((.((((......(((((((.....))))))).)))))))..)))))))).... ( -40.70, z-score = -4.41, R) >droEre2.scaffold_4770 3326035 115 + 17746568 CGGAUAUAGACACGAGCACGGCGAGAUACGAGCUACGGAUACG--UACUAGUGUUCCGACCAGGCACGCAAUGGAAAUGGCCAGAAAAUGGCCAUAUGUGGCCAAGUCGGCUCCCAC .............((((.(((((..((((.((.((((....))--)))).))))..).....(((.((((......(((((((.....))))))).)))))))..)))))))).... ( -40.70, z-score = -2.77, R) >droAna3.scaffold_13340 13064855 101 + 23697760 UAUAUAUAGACUAGAAUAC----AGAUAC-AGAUACGGAUACGGAUA--------CGGGCCAGGCACGCAAUGG---CAGCCAGAAAAUGGCCAUAUGUGGCCAAGUCGGCUCCCAC ...................----......-.....((....))....--------.(((((.(((..((....)---).))).((...((((((....))))))..))))))).... ( -26.30, z-score = -1.09, R) >consensus CGGAUAUAGACACGAGCACGGCGAGAUAC_AGAUACGGAUACG_AUACUAGUAUUCCGACCAGGCACGCAAUGGAAAUGGCCAGAAAAUGGCCAUAUGUGGCCAAGUCGGCUCCCAC .............((((.((((.............(((((((........))).))))....(((.((((......(((((((.....))))))).)))))))..)))))))).... (-30.29 = -31.93 + 1.64)
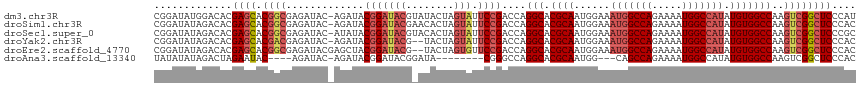
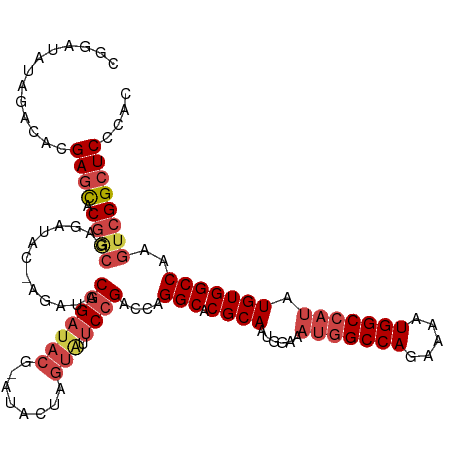
| Location | 7,193,152 – 7,193,264 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.28 |
| Shannon entropy | 0.37038 |
| G+C content | 0.49442 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -17.01 |
| Energy contribution | -19.10 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7193152 112 - 27905053 GCACACACUG-----CCGUACAUUGCAGCCUAGGAUACUACGGAUAUGGACACGAGCACGGCGAGAUAC-AGAUACGGAUACGUAUACUAGUAUUCCGACCAGGCACGCAAUGGAAAU (((.....))-----)....((((((..(((((....))).))............((..((((..((((-(((((((....))))).)).))))..)).))..))..))))))..... ( -31.20, z-score = -2.65, R) >droSim1.chr3R 13356238 112 + 27517382 GCACACACUG-----CCGUACAUUGCAGCCUGGGAUACUACGGAUAUAGACACGAGCACGGCGAGAUAC-AGAUACGGAUACGAACACUAGUAUUCCGACCAGGCACGCAAUGGAAAU (((.....))-----)....((((((.((((((....(....)..........................-.....(((((((........))).)))).))))))..))))))..... ( -28.90, z-score = -2.55, R) >droSec1.super_0 6358815 112 + 21120651 GCACAUACUG-----UCGUACAUUGCAGCCUGGGAUACUACGGAUAUAGACACGAGCACGGCGAGAUAC-AUAUACGGAUACGUACACUAGUAUUCCGACCAGGCACGCAAUGGAAAU ..........-----.....((((((.((((((....(....)...............(((...(((((-...((((....)))).....)))))))).))))))..))))))..... ( -29.10, z-score = -1.84, R) >droYak2.chr3R 11235111 115 - 28832112 GCACACACUGCCGUACCGUACAUUGCAGCUAGGGAUACUACGGAUAUAGACACGAGCACGACGAGAUAC-AGAUACGGAUACGUA--CUAGUAUUCCGACCAGGCACGCAAUGGAAAU (((.....))).........((((((.(((.((....(....)..................((..((((-((.((((....))))--)).))))..)).)).)))..))))))..... ( -26.40, z-score = -1.24, R) >droEre2.scaffold_4770 3326074 111 + 17746568 GCACACACUG-----CCGUACAUUGCAGCCUGGGAUACUACGGAUAUAGACACGAGCACGGCGAGAUACGAGCUACGGAUACGUA--CUAGUGUUCCGACCAGGCACGCAAUGGAAAU (((.....))-----)....((((((.((((((....(....)..........((((((....((.((((..(....)...))))--)).))))))...))))))..))))))..... ( -34.80, z-score = -2.60, R) >droAna3.scaffold_13340 13064893 87 + 23697760 -----------------GCACA-GGCACACAAACAUAUAGCAUAUAUAUAGACUAGAAUAC--AGAUAC-AGAUACGGAUACGGA--------UACGGGCCAGGCACGCAAUGGCA-- -----------------((...-.((........(((((.....)))))............--......-......((...((..--------..))..))......))....)).-- ( -11.00, z-score = -0.60, R) >consensus GCACACACUG_____CCGUACAUUGCAGCCUGGGAUACUACGGAUAUAGACACGAGCACGGCGAGAUAC_AGAUACGGAUACGUA__CUAGUAUUCCGACCAGGCACGCAAUGGAAAU ....................((((((.((((((....(....)................................(((((((........)))).))).))))))..))))))..... (-17.01 = -19.10 + 2.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:06 2011