
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,192,603 – 7,192,711 |
| Length | 108 |
| Max. P | 0.841907 |

| Location | 7,192,603 – 7,192,711 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.93 |
| Shannon entropy | 0.53230 |
| G+C content | 0.43924 |
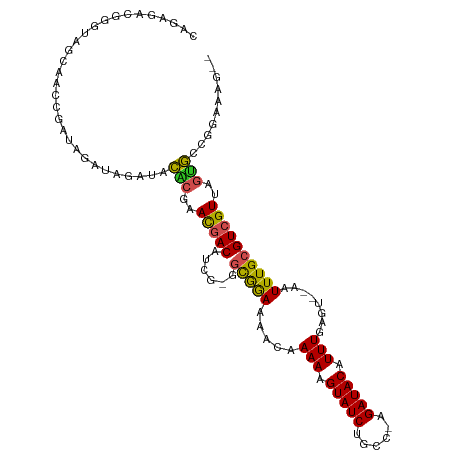
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -12.10 |
| Energy contribution | -11.28 |
| Covariance contribution | -0.82 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.841907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
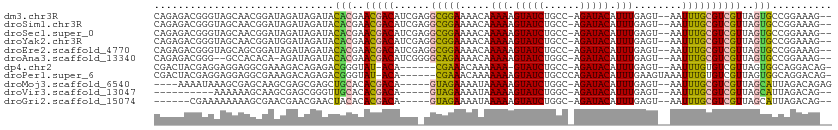
>dm3.chr3R 7192603 108 + 27905053 CAGAGACGGGUAGCAACGGAUAGAUAGAUACACGAACGACAUCGAGGCGGAAAACAAAAAGUAUCUGCC-AGAUACAUUUGAGU--AAUUUGCGUCGUUAGUGCCGGAAAG-- ......(.((((..(((((...(.(((((((.((((.(..(((..((((((..((.....)).))))))-.))).).)))).))--.)))))).)))))..)))).)....-- ( -24.30, z-score = -1.44, R) >droSim1.chr3R 13355685 108 - 27517382 CAGAGACGGGUAGCAACGGAUAGAUAGAUACACGAACGACAUCGAGGCGGAAAACAAAAAGUAUCUGCC-AGAUACAUUUGAGU--AAUUUGCGUCGUUAGUGCCGGAAAG-- ......(.((((..(((((...(.(((((((.((((.(..(((..((((((..((.....)).))))))-.))).).)))).))--.)))))).)))))..)))).)....-- ( -24.30, z-score = -1.44, R) >droSec1.super_0 6358256 108 - 21120651 CAGAGACGGGUAGCAACGGAUAGAUAGAUACACGAACGACAUCGAGGCGGAAAACAAAAAGUAUCUGCC-AGAUACAUUUGAGU--AAUUUGCGUCGUUAGUGCCGGAAAG-- ......(.((((..(((((...(.(((((((.((((.(..(((..((((((..((.....)).))))))-.))).).)))).))--.)))))).)))))..)))).)....-- ( -24.30, z-score = -1.44, R) >droYak2.chr3R 11234533 108 + 28832112 CAGAGACGGGUAGCAACGGAUGGAUAGAUACACGAACGACAUCGAGGCGGAAAACAAAAAGUAUCUGCC-AGAUACAUUUGAGU--AAUUUGCGUCGUUAGUGCCGGAAAG-- ......(.((((..(((.((((.................))))((.(((((..((..((((((((....-.))))).)))..))--..))))).)))))..)))).)....-- ( -24.43, z-score = -1.10, R) >droEre2.scaffold_4770 3325534 108 - 17746568 CAGAGACGGGUAGCAGCGGAUAGAUAGAUACACGAACGACAUCGAGGCGGAAAACAAAAAGUAUCUGCC-AGAUACAUUUGAGU--AAUUUGCGUCGUUAGUGCCGGAAAG-- ......(.((((..(((((...(.(((((((.((((.(..(((..((((((..((.....)).))))))-.))).).)))).))--.)))))).)))))..)))).)....-- ( -24.30, z-score = -1.20, R) >droAna3.scaffold_13340 13064339 105 - 23697760 CAGAGACGGG--GCCACACA-AGAUAGAUACACGAACGACAUCGGGGCAGAAAACAAAAAGUAUCUGGC-AGAUACAUUUGAGU--AAUUUGCGUCGUUAGUGCCGGAAAG-- ......((((--......).-.........(((.((((((.......((((..((.....)).))))((-((((((......))--.)))))))))))).)))))).....-- ( -23.00, z-score = -1.38, R) >dp4.chr2 1572863 101 - 30794189 CGACUACGAGGAGGAGGCGAAAGACAGAGACGGGUAU-ACA------CGAAACAAAAAA-GUAUCUGCC-AGAUACAUUUGAGU--AAUUUGUGUCGUUAGUGGCAGGACAG- (..((((...((.((.(((((..((.....((.....-...------))...((((...-(((((....-.))))).)))).))--..))))).)).)).))))..).....- ( -20.80, z-score = -0.99, R) >droPer1.super_6 1597473 105 - 6141320 CGACUACGAGGAGGAGGCGAAAGACAGAGACGGGUAU-ACA------CGAAACAAAAAAAGUAUCUGCCCAGAUACAUUUGAAGUAAAUUUGUGUCGUUAGUGGCAGGACAG- (..((((...((.((.(((((..((.....((.....-...------))...........((((((....)))))).......))...))))).)).)).))))..).....- ( -21.30, z-score = -0.94, R) >droMoj3.scaffold_6540 13849250 101 - 34148556 ----AAAAUAAAGCGAGCAAGCGAGCGAGCUGCACACGACA-----GUAGAAAAUAAAAAGUAUCUGGC-AGAUACAUUUGAGU--AAUUUGCGUCGUUAGCAUUAGACAGAG ----.......(((..((......))..)))((..(((((.-----(((((.....(((.(((((....-.))))).)))....--..))))))))))..))........... ( -22.30, z-score = -1.16, R) >droVir3.scaffold_13047 15964759 93 - 19223366 ----------AAAAAAGCAAGCGAGCGGGUUGCACACGACA-----GUAGAAAAUAAAAAGUAUCUGGC-AGAUACAUUUGAGU--AAUUUGCGUCGUUAGCAUUAGACAG-- ----------......((.((((((((((((((..((....-----))............(((((....-.)))))......))--))))))).))))).)).........-- ( -23.60, z-score = -2.78, R) >droGri2.scaffold_15074 110666 97 - 7742996 ------CGAAAAAAAAGCGAACGAACGAACUACACACGACA-----GUAGAAAAUAAAAAGUAUCUGGC-AGAUACAUUUGAGU--AAUUUGCGUCGUUAGCAUUAGACAG-- ------..........((.(((((.((..((((........-----))))..........(((((....-.)))))........--......))))))).)).........-- ( -17.70, z-score = -1.54, R) >consensus CAGAGACGGGUAGCAACCGAUAGAUAGAUACACGAACGACAUCG_GGCGGAAAACAAAAAGUAUCUGCC_AGAUACAUUUGAGU__AAUUUGCGUCGUUAGUGCCGGAAAG__ ..............................(((..(((((......(((((.....(((.((((((....)))))).)))........))))))))))..))).......... (-12.10 = -11.28 + -0.82)
| Location | 7,192,603 – 7,192,711 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.93 |
| Shannon entropy | 0.53230 |
| G+C content | 0.43924 |
| Mean single sequence MFE | -17.77 |
| Consensus MFE | -9.89 |
| Energy contribution | -9.81 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7192603 108 - 27905053 --CUUUCCGGCACUAACGACGCAAAUU--ACUCAAAUGUAUCU-GGCAGAUACUUUUUGUUUUCCGCCUCGAUGUCGUUCGUGUAUCUAUCUAUCCGUUGCUACCCGUCUCUG --.......((((.((((((((....(--((......)))...-(((.((.((.....))..)).)))..).))))))).))))............................. ( -20.10, z-score = -1.31, R) >droSim1.chr3R 13355685 108 + 27517382 --CUUUCCGGCACUAACGACGCAAAUU--ACUCAAAUGUAUCU-GGCAGAUACUUUUUGUUUUCCGCCUCGAUGUCGUUCGUGUAUCUAUCUAUCCGUUGCUACCCGUCUCUG --.......((((.((((((((....(--((......)))...-(((.((.((.....))..)).)))..).))))))).))))............................. ( -20.10, z-score = -1.31, R) >droSec1.super_0 6358256 108 + 21120651 --CUUUCCGGCACUAACGACGCAAAUU--ACUCAAAUGUAUCU-GGCAGAUACUUUUUGUUUUCCGCCUCGAUGUCGUUCGUGUAUCUAUCUAUCCGUUGCUACCCGUCUCUG --.......((((.((((((((....(--((......)))...-(((.((.((.....))..)).)))..).))))))).))))............................. ( -20.10, z-score = -1.31, R) >droYak2.chr3R 11234533 108 - 28832112 --CUUUCCGGCACUAACGACGCAAAUU--ACUCAAAUGUAUCU-GGCAGAUACUUUUUGUUUUCCGCCUCGAUGUCGUUCGUGUAUCUAUCCAUCCGUUGCUACCCGUCUCUG --.......((((.((((((((....(--((......)))...-(((.((.((.....))..)).)))..).))))))).))))............................. ( -20.10, z-score = -1.21, R) >droEre2.scaffold_4770 3325534 108 + 17746568 --CUUUCCGGCACUAACGACGCAAAUU--ACUCAAAUGUAUCU-GGCAGAUACUUUUUGUUUUCCGCCUCGAUGUCGUUCGUGUAUCUAUCUAUCCGCUGCUACCCGUCUCUG --.......((((.((((((((....(--((......)))...-(((.((.((.....))..)).)))..).))))))).))))............................. ( -20.10, z-score = -1.27, R) >droAna3.scaffold_13340 13064339 105 + 23697760 --CUUUCCGGCACUAACGACGCAAAUU--ACUCAAAUGUAUCU-GCCAGAUACUUUUUGUUUUCUGCCCCGAUGUCGUUCGUGUAUCUAUCU-UGUGUGGC--CCCGUCUCUG --.......((((.(((((((((...(--((......)))..)-))((((.((.....))..)))).......)))))).))))........-........--.......... ( -20.40, z-score = -1.43, R) >dp4.chr2 1572863 101 + 30794189 -CUGUCCUGCCACUAACGACACAAAUU--ACUCAAAUGUAUCU-GGCAGAUAC-UUUUUUGUUUCG------UGU-AUACCCGUCUCUGUCUUUCGCCUCCUCCUCGUAGUCG -...............((((((....(--((......)))...-((((((.((-............------...-......)).))))))...............)).)))) ( -12.20, z-score = 0.06, R) >droPer1.super_6 1597473 105 + 6141320 -CUGUCCUGCCACUAACGACACAAAUUUACUUCAAAUGUAUCUGGGCAGAUACUUUUUUUGUUUCG------UGU-AUACCCGUCUCUGUCUUUCGCCUCCUCCUCGUAGUCG -.....((((.....((((.(((((........(((.((((((....)))))).)))))))).)))------)..-.....((...........))..........))))... ( -12.60, z-score = 0.50, R) >droMoj3.scaffold_6540 13849250 101 + 34148556 CUCUGUCUAAUGCUAACGACGCAAAUU--ACUCAAAUGUAUCU-GCCAGAUACUUUUUAUUUUCUAC-----UGUCGUGUGCAGCUCGCUCGCUUGCUCGCUUUAUUUU---- ..........(((..((((((......--........(((((.-....)))))..............-----))))))..)))((..((......))..))........---- ( -17.69, z-score = -0.73, R) >droVir3.scaffold_13047 15964759 93 + 19223366 --CUGUCUAAUGCUAACGACGCAAAUU--ACUCAAAUGUAUCU-GCCAGAUACUUUUUAUUUUCUAC-----UGUCGUGUGCAACCCGCUCGCUUGCUUUUUU---------- --........(((..((((((......--........(((((.-....)))))..............-----))))))..)))....((......))......---------- ( -15.79, z-score = -1.45, R) >droGri2.scaffold_15074 110666 97 + 7742996 --CUGUCUAAUGCUAACGACGCAAAUU--ACUCAAAUGUAUCU-GCCAGAUACUUUUUAUUUUCUAC-----UGUCGUGUGUAGUUCGUUCGUUCGCUUUUUUUUCG------ --.........((.(((((.((.((((--((.((...(((((.-....)))))..............-----.....)).)))))).))))))).))..........------ ( -16.25, z-score = -1.22, R) >consensus __CUUUCCGGCACUAACGACGCAAAUU__ACUCAAAUGUAUCU_GGCAGAUACUUUUUGUUUUCCGCC_CGAUGUCGUUCGUGUAUCUAUCUAUCCGUUGCUACCCGUCUCUG ..........(((..((((((............(((.((((((....)))))).)))...............))))))..))).............................. ( -9.89 = -9.81 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:06:04 2011