| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,159,050 – 7,159,164 |
| Length | 114 |
| Max. P | 0.654484 |

| Location | 7,159,050 – 7,159,164 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.01 |
| Shannon entropy | 0.57801 |
| G+C content | 0.58766 |
| Mean single sequence MFE | -41.55 |
| Consensus MFE | -18.50 |
| Energy contribution | -20.11 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
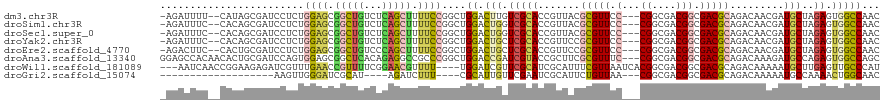
>dm3.chr3R 7159050 114 + 27905053 -AGAUUUU--CAUAGCGAUCCUCUGGAGCGGCUGUCUCAGCUUUUCCGGCUGGACUUGUCGCACCGUUACGCGUUCC---CGGCGACGGCGACGCAGACAACGAUGCUAGAGUGGCCAAC -.......--......(..(((((((.(((((.(((.(((((.....))))))))..)))))..((((..(((((.(---((....))).)))))....))))...)))))).)..)... ( -43.70, z-score = -1.57, R) >droSim1.chr3R 13322415 114 - 27517382 -AGAUUUC--CACAGCGAUCCUCUGGAGCGGCUGUCUCAGCUUUUCCGGCUGGACUGGUCGCACCGUUACGCGUUCC---CGGCGACGGCGACGCAGACAACGAUGCUAGAGUGGCCAAC -......(--((((((.(((....((.(((((((((.(((((.....)))))))).)))))).))(((..(((((.(---((....))).))))).)))...))))))...))))..... ( -45.30, z-score = -1.44, R) >droSec1.super_0 6324994 114 - 21120651 -AGAUUUC--CACAGCGAUCCUCUGGAGCGGCUGUCUCAGCUUUUCCGGCUGGACUGGUCGCACCGUUACGCGUUCC---CGGCGACGGCGACGCAGACAACGAUGCUAGAGUGGCCAAC -......(--((((((.(((....((.(((((((((.(((((.....)))))))).)))))).))(((..(((((.(---((....))).))))).)))...))))))...))))..... ( -45.30, z-score = -1.44, R) >droYak2.chr3R 11195553 114 + 28832112 -AGAUUUC--CACAGCGAUCCUCUGGAGCGGCUGUCUCAGCUUUUCCGGCUGGACUGCUCGCACCGUUCCGCGUUCC---CGGCGACGGCGACGCAGACAACGAUGCUAGAGUGGCCAAC -......(--(((((((.(((.((((((.(((((...))))).))))))..))).)))).(((.(((((.(((((.(---((....))).))))).))..))).)))....))))..... ( -47.70, z-score = -2.05, R) >droEre2.scaffold_4770 3286809 114 - 17746568 -AGACUUC--CACUGCGAUCCUCUGGAGCGGCUGUCCCAGCUUUUCCGGCUGGACUGCUCGCACCGUUCCGCGUUCC---CGGCGACGGCGACGCAGACAACGAUGCUAGAGUGGCCAAC -......(--(((((((.(((.((((((.(((((...))))).))))))..))).)))..(((.(((((.(((((.(---((....))).))))).))..))).)))...)))))..... ( -47.70, z-score = -2.00, R) >droAna3.scaffold_13340 13031878 117 - 23697760 GGAGCCACAACACUGCGAUCCAGUGGAGCGGCUCACAGAGGCCGCCCGGCUGGACCGAUCGUACCGCUUCGCGUUUC---CGGCGACGGCGACGCAGACAAAGAUGCCAGAGUGGCCAGC .(((((.(..(((((.....)))))..).))))).....(((((((.(((.(((.((.......)).)))(((((.(---((....))).)))))..........))).).))))))... ( -51.00, z-score = -2.01, R) >droWil1.scaffold_181089 8050797 113 - 12369635 ---AAUCAACCGGAAGAGAUCGUUUGAACCGUUUUCGGAACGUUUU----UGGAUCGUUCGCAUCGCAUUUCGUUAAUCACGGCGACGGCGACGCAGACAAAAAUGCUUGAGUUGCCCAU ---..(((((((((((...((....))....)))))))..((((((----((((....))((.((((...(((((......)))))..)))).))...)))))))).))))......... ( -27.50, z-score = 1.13, R) >droGri2.scaffold_15074 68397 90 - 7742996 -------------------AAGUUGGGAUCGCAU----AGAUCUUU----CGCAUUGUUCGAAUCGCAUUCUGUUAA---CGGCGACGGCGACGCAGACAAAAAUGCCAAAACUGGCAAC -------------------..((.((((((....----.)))))).----.)).((((..(..((((...(((....---))).....))))..)..))))...(((((....))))).. ( -24.20, z-score = -0.87, R) >consensus _AGAUUUC__CACAGCGAUCCUCUGGAGCGGCUGUCUCAGCUUUUCCGGCUGGACUGGUCGCACCGUUACGCGUUCC___CGGCGACGGCGACGCAGACAACGAUGCUAGAGUGGCCAAC ..............(((((((..(((((.(((((...))))).)))))...)))....))))...................(((.(((....)(((........)))....)).)))... (-18.50 = -20.11 + 1.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:59 2011