| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,010,367 – 7,010,502 |
| Length | 135 |
| Max. P | 0.998210 |

| Location | 7,010,367 – 7,010,469 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.93 |
| Shannon entropy | 0.26324 |
| G+C content | 0.46209 |
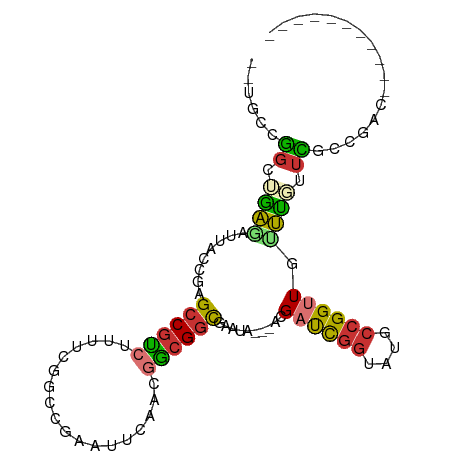
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -24.86 |
| Energy contribution | -25.58 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.995970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
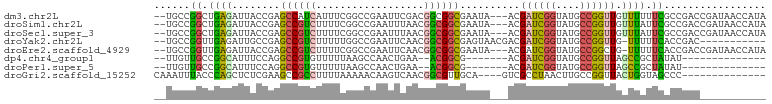
>dm3.chr2L 7010367 102 - 23011544 CGAAUUCGACGGCGGCGAAUA---ACGAUCGGUAUGCCGGUUGUUUUUUCGCCGACCGAUAACCAUAGAAAUGUGAAAAAAACCUAAGAAAUACGAAGCUUUUCG ((((((((.((((((((((.(---((((((((....)))))))))..))))))).))).....((((....))))..................))))....)))) ( -37.20, z-score = -4.65, R) >droSim1.chr2L 6799849 101 - 22036055 CGAAUUUAACGGCGGCGAAUA---ACGAUCGGUAUGCCGGUUGUUUAUUCGCCGACCGAUAACCAUACUAAUGUGAAAAAAACCUAAGAAAUCGGGAGCUUUCG- ((((.....((((((((((((---((((((((....))))))).)))))))))).)))........................((..(....)..))....))))- ( -36.20, z-score = -4.29, R) >droSec1.super_3 2536157 100 - 7220098 CGAAUUUAACGGCGGCGAAUA---ACGAUCGGUAUGCCGGUUGUUUAUUCGCCGACCGAUAACCAUACUAAUGUG-AAAAAACCUAAGAAAUCGGGAGCUUUCG- .........((((((((((((---((((((((....))))))).)))))))))).)))................(-(((...((..(....)..))...)))).- ( -36.40, z-score = -4.30, R) >droYak2.chr2L 16418367 90 + 22324452 CGAAUUCAACGGCGGCGAGUAACGACGAUCGGUAUGCCGGUUGUUU-UUCACCGACU------------UGUGUG-AAGAAACCUAAGAAAUCGGAAGCUUUCG- ....(((((((((((.(((....(((((((((....))))))))).-))).))).).------------))).))-)).........((((.(....).)))).- ( -25.70, z-score = -0.75, R) >droEre2.scaffold_4929 15919275 99 - 26641161 CGAAUUCAACGGCGGCGAAUA---ACGAUCGGUAUGCCGGCUGUUU-UUCACCGACCGAUAACCAUACUAAUGUG-AAAAAACCCAAGACAUCGGGAGCUUUCG- ((((((((.((((((.(((.(---(((..(((....)))..)))).-))).))).))).....(((....)))))-))....(((........)))....))))- ( -28.80, z-score = -2.10, R) >consensus CGAAUUCAACGGCGGCGAAUA___ACGAUCGGUAUGCCGGUUGUUU_UUCGCCGACCGAUAACCAUACUAAUGUG_AAAAAACCUAAGAAAUCGGGAGCUUUCG_ .........((((((((((.....((((((((....))))))))...))))))).))).............................((((.(....).)))).. (-24.86 = -25.58 + 0.72)
| Location | 7,010,405 – 7,010,502 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 63.42 |
| Shannon entropy | 0.67358 |
| G+C content | 0.52832 |
| Mean single sequence MFE | -33.51 |
| Consensus MFE | -14.78 |
| Energy contribution | -14.17 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.72 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
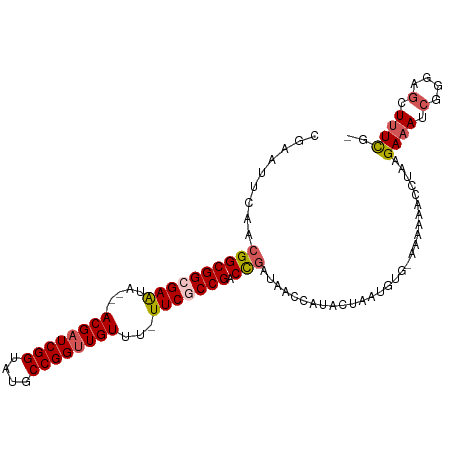
>dm3.chr2L 7010405 97 - 23011544 --UGCCGGCUGAGAUUACCGAGCCAUCAUUUCGGCCGAAUUCGACGGCGGCGAAUA---ACGAUCGGUAUGCCGGUUGUUUUUUCGCCGACCGAUAACCAUA --...((((((((((....((....)))))))))))).......((((((((((.(---((((((((....)))))))))..))))))).)))......... ( -43.20, z-score = -4.34, R) >droSim1.chr2L 6799886 97 - 22036055 --UGCCGGCUGAGAUUACCGAGCCGUCUUUUCGGCCGAAUUUAACGGCGGCGAAUA---ACGAUCGGUAUGCCGGUUGUUUAUUCGCCGACCGAUAACCAUA --...(((((((((..((......))..))))))))).......((((((((((((---((((((((....))))))).)))))))))).)))......... ( -45.90, z-score = -5.27, R) >droSec1.super_3 2536193 97 - 7220098 --UGCCGGCUGAGAUUACCGAGCCGUCUUUUCGGCCGAAUUUAACGGCGGCGAAUA---ACGAUCGGUAUGCCGGUUGUUUAUUCGCCGACCGAUAACCAUA --...(((((((((..((......))..))))))))).......((((((((((((---((((((((....))))))).)))))))))).)))......... ( -45.90, z-score = -5.27, R) >droYak2.chr2L 16418402 88 + 22324452 --UGCCGGUUGAGAUUGCCGAGCCGUCUUUUUGGCCGAAUUCAACGGCGGCGAGUAACGACGAUCGGUAUGCCGGUUG-UUUUUCACCGAC----------- --.((((.(((((.((((((((.......))))).))).)))))))))((.(((....(((((((((....)))))))-)).))).))...----------- ( -32.80, z-score = -1.61, R) >droEre2.scaffold_4929 15919311 96 - 26641161 --UGCCGGUUGAGAUUACCGAGCCGUCUUUUCGGCCGAAUUCAACGGCGGCGAAUA---ACGAUCGGUAUGCCGGCUG-UUUUUCACCGACCGAUAACCAUA --....(((((.((....((.((((......))))))...))..((((((.(((.(---(((..(((....)))..))-)).))).))).))).)))))... ( -36.50, z-score = -2.76, R) >dp4.chr4_group1 1556336 77 + 5278887 --UUGUUGCCGGCAUUUCCAGGCCGUGUUUUUAAGCCAACUGAA--ACGGCG-------ACGAUCGGUAUGCCGGUUAGCCGCUAUAU-------------- --..(((((((((((..((..(((((..................--)))))(-------.....))).))))))).))))........-------------- ( -23.67, z-score = -0.60, R) >droPer1.super_5 1553676 77 - 6813705 --UUGUUGCCGGCAUUUCCAGGCCGUGUUUUUAAGCCAACUGAA--ACGGCG-------ACGAUCGGUAUGCCGGUUAGCCGCUAUAU-------------- --..(((((((((((..((..(((((..................--)))))(-------.....))).))))))).))))........-------------- ( -23.67, z-score = -0.60, R) >droGri2.scaffold_15252 3011254 84 - 17193109 CAAAUUUACCCAGCUCUCGAAGCCGCCUUUUAAAAACAAGUCAACGGCGUUGCA----GUCGCCUAACUUGCCGGUUACUGGUAGCCC-------------- ............(((((.(.(((((...........(((((....((((.....----..))))..))))).))))).).)).)))..-------------- ( -16.40, z-score = 0.60, R) >consensus __UGCCGGCUGAGAUUACCGAGCCGUCUUUUCGGCCGAAUUCAACGGCGGCGAAUA___ACGAUCGGUAUGCCGGUUGUUUGUUCGCCGAC___________ ......((.((((........((((((..................))))))..........((((((....)))))).)))).))................. (-14.78 = -14.17 + -0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:46 2011