| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,140,669 – 7,140,764 |
| Length | 95 |
| Max. P | 0.989782 |

| Location | 7,140,669 – 7,140,764 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.17 |
| Shannon entropy | 0.49898 |
| G+C content | 0.45798 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -13.55 |
| Energy contribution | -14.07 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989782 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
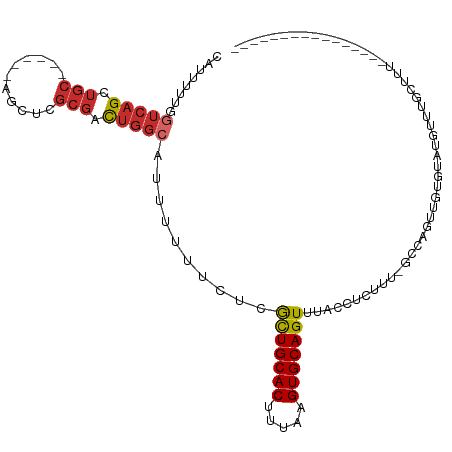
>dm3.chr3R 7140669 95 + 27905053 CAUUUUUGGUCAGCUGC------AGUUCGCGACUGGCAUUUUUUCUCGCUGCACUUUAAGUGCAGUUUACCUCUUU-GGCAGUUGUGUAUGUUUGCUAUGCA------------- ...........(((.((------(...((((((((.((.........(((((((.....))))))).........)-).))))))))..)))..))).....------------- ( -31.27, z-score = -2.73, R) >droSim1.chr3R 13301880 95 - 27517382 CAUUUUUGGUCAGCUGC------AGCUCGCGACUGGCAUUUUUUCUCGCUGCACUUUAAGUGCAGUUUACCUCUUU-GGCAGUUGUGCAUGUUUGCUUUGCA------------- ............((.((------(((.((((((((.((.........(((((((.....))))))).........)-).))))))))...).))))...)).------------- ( -30.67, z-score = -1.72, R) >droEre2.scaffold_4770 3268042 95 - 17746568 CAUUUUUGGUCAGCUGC------AGCUCGCGACUGGCAUUUUUUCUCGCUGCACUUUAAGUGCAGUUUACCUCUUU-GGCAGUUGUGUAUGUUUGCUUUCCA------------- ......(((..(((.((------(...((((((((.((.........(((((((.....))))))).........)-).))))))))..)))..)))..)))------------- ( -34.77, z-score = -4.39, R) >droAna3.scaffold_13340 13012129 115 - 23697760 CAUUUUCGGUGAGCUGCUGCACCAACUCGCGACGGGCAUUUUUUCUCGCUGCACUUUAAGUGCAGUUUACCUCUUUUGCCAGUUGCAUGAAUGAGUGUGUGUGUUUGCUUUUCCC .......((.((((....(((((((((((((((.((((.........(((((((.....)))))))..........)))).)))))......)))).)).))))..))))..)). ( -37.51, z-score = -2.62, R) >dp4.chr2 1523853 74 - 30794189 CAUUUUUGGUCAGC----------GCCCGCGACUGGCAUUUUUUCUCGCUGCACUUUAAGUGCAGUUUACCUCUUG-GCCAUUUG------------------------------ ......(((((((.----------(((.......)))..........(((((((.....))))))).......)))-))))....------------------------------ ( -21.90, z-score = -2.57, R) >droWil1.scaffold_181089 8029837 93 - 12369635 CAUUUUUGGUCAGUUGC------AGCUCGCGACUGGAAUUUUUUCUCAUUGCACUUUAAGUGCAGUUUACCUCUUU-GGCAGGC-UCUAUGUUUGCUUUGG-------------- .......((((((((((------.....)))))))............(((((((.....)))))))..))).((..-(((((((-.....)))))))..))-------------- ( -28.20, z-score = -2.82, R) >droVir3.scaffold_13047 15883394 93 - 19223366 CAUUUUUGGUCAGUUGC------AGUUCGCGACUGGCAUUUUUUCUCGCUGCACUUUAAGUGCAGUUUAGCCUGUA-CCCAGCAGCCAACCCCAAACCCA--------------- ....(((((((((((((------.....)))))))))..........(((((.......((((((......)))))-)...)))))......))))....--------------- ( -28.30, z-score = -3.29, R) >droMoj3.scaffold_6540 13786842 89 - 34148556 CAUUUUUGGUCAGCUGC------AGAUCGCGAUUGCAAUUUUUUCUCGCUGCACUUUAAGUGCAGUUUACCUCUUU-GCCAUGUUUGUGCCCUCAG------------------- .............(((.------((..(((((..((((.........(((((((.....)))))))........))-)).....)))))..)))))------------------- ( -19.63, z-score = -0.60, R) >droGri2.scaffold_15074 45115 80 - 7742996 CAUUUUUGGUCAGUUGC------AGCUCGCGACUGGCAUUUUUUCUCGUUGCACUUUAAGUGCAGUUUACCUCUUU-GCCAUAUUUG---------------------------- ........(((((((((------.....)))))))))...........((((((.....))))))...........-..........---------------------------- ( -20.90, z-score = -2.64, R) >consensus CAUUUUUGGUCAGCUGC______AGCUCGCGACUGGCAUUUUUUCUCGCUGCACUUUAAGUGCAGUUUACCUCUUU_GCCAGUUGUGUAUGUUUGCUUU________________ ........(((((.(((...........))).)))))..........(((((((.....)))))))................................................. (-13.55 = -14.07 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:54 2011