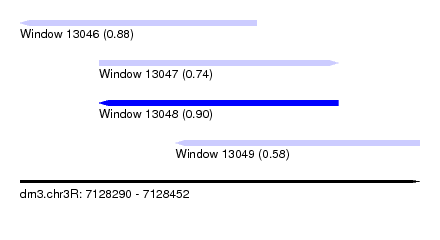
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,128,290 – 7,128,452 |
| Length | 162 |
| Max. P | 0.900954 |

| Location | 7,128,290 – 7,128,386 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 62.05 |
| Shannon entropy | 0.74730 |
| G+C content | 0.39937 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -9.08 |
| Energy contribution | -10.03 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
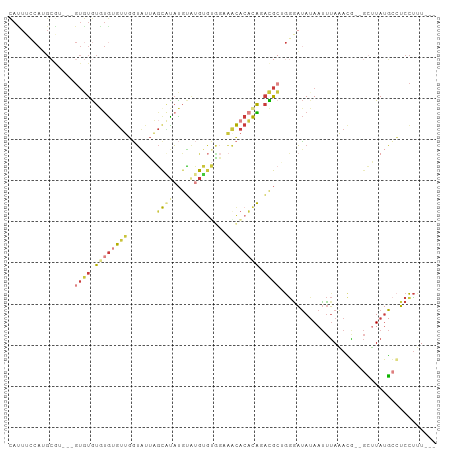
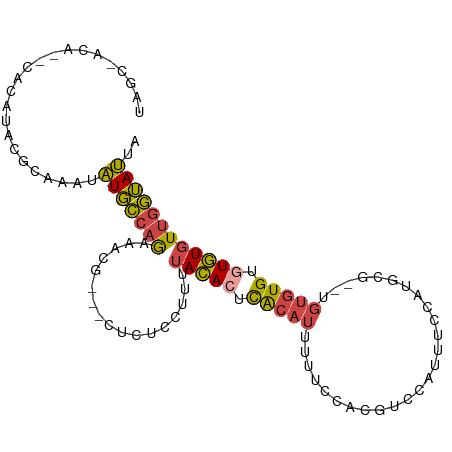
>dm3.chr3R 7128290 96 - 27905053 CAUUUCCAUGCGU---GUGUGUGUGUGUUGGUAUUAGCAUAUGUAUGUGUGGAAACACACAGACGCUGGGAUAUAAUUUAAACG--GCUUAUGCCUCCUUU--- ...(((((.((((---.((((..(((((((....)))))))..)))(((((....)))))).)))))))))............(--((....)))......--- ( -33.70, z-score = -3.12, R) >droEre2.scaffold_4770 3255756 96 + 17746568 CAUUUCCAUGCAU---GUGUGUAUGUGUUGGUAUUAGCUUAUGUAUGUGUAGAAGCACACAUACGCAGGAAUAAAAUUUAAACG--GCUUAUGCCUCCUUU--- ...((((.(((.(---(((((((((..(.(((....))).)..)))((......))))))))).)))))))............(--((....)))......--- ( -23.10, z-score = -0.51, R) >droYak2.chr3R 11164173 96 - 28832112 CACUUUCAAGCGA---GUGUGUGUGUGUUGGUAUUAGCAUAUGUAUGUGCAGAAGCACACAGACGCAGGGAUAUAAUUUAAACA--GCUUAUGGCUCCUUU--- .............---((((.((((((((.((((..((....))..))))...)))))))).))))((((.(((((........--..)))))..))))..--- ( -24.50, z-score = -0.15, R) >droSec1.super_0 6294190 98 + 21120651 CAUUUCCAUGCGU-CUGUGUGUGUGUGUUGGUAUUAGCAUAUGUGUGUGUGGCAACACACAGACGCUGGGAUAUAAUUUAAACG--GCUUAUGCCUCCUUU--- ...(((((.((((-((((((((((((((((....)))))))).(((.....))))))))))))))))))))............(--((....)))......--- ( -37.30, z-score = -3.55, R) >droSim1.chr3R 13286011 98 + 27517382 CAUUUCCAUGCGU-CUGUGUGUGUGUGUUGGUAUUAGCAUAUGUGUGUGUGGCAACACACAGACGCUGGGAUAUAAUUUAAACG--GCUUAUGCCUCCUUU--- ...(((((.((((-((((((((((((((((....)))))))).(((.....))))))))))))))))))))............(--((....)))......--- ( -37.30, z-score = -3.55, R) >dp4.chr2 1509858 84 + 30794189 -----------------AUUGUACGAGUAUAUGCCUGUGCACGCACACACGCACACACACGCAUAAAAGGAUAUAAUUUAUAUGCCGUGUAGGCGUCUCUU--- -----------------.......(((...((((((((((..........))))..(((((((((.(((.......))).)))).))))))))))))))..--- ( -22.40, z-score = -0.95, R) >droAna3.scaffold_13340 12999887 104 + 23697760 UAUUAACAAGGACACUGUAUAUAUAAAAAGUACCUACCUUUUUGCCGAAUAAAUCUGAUUGAUUGAUGAGUAAUAACCAAUAAUAAAAAUAUUAAGCCUAUCUG ........(((.....((((...(((((((.......))))))).......((((.....))))........................))))....)))..... ( -7.20, z-score = 0.99, R) >consensus CAUUUCCAUGCGU___GUGUGUGUGUGUUGGUAUUAGCAUAUGUAUGUGUGGAAACACACAGACGCUGGGAUAUAAUUUAAACG__GCUUAUGCCUCCUUU___ ................((((.((((((((.......(((((....)))))...)))))))).))))...................................... ( -9.08 = -10.03 + 0.95)


| Location | 7,128,322 – 7,128,419 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 72.15 |
| Shannon entropy | 0.51535 |
| G+C content | 0.42144 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -14.87 |
| Energy contribution | -14.85 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
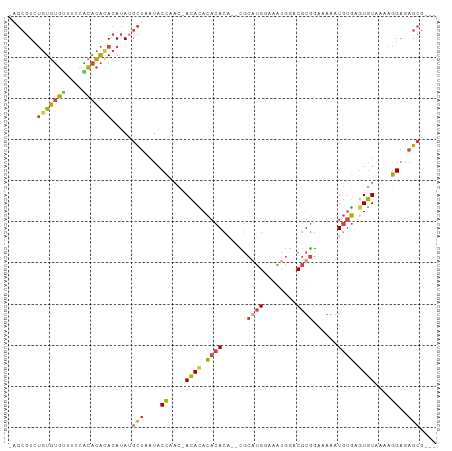
>dm3.chr3R 7128322 97 + 27905053 -AGCGUCUGUGUGUUUCCACACAUACAUAUGCUAAUACCAAC-ACACACACACA--CGCAUGGAAAUGGACGUGGAAAAAUGUGAGUGUAAAAGGAGAGCG--- -(((((.(((((((......))))))).)))))....((.((-((.((((..((--(((((....)))..))))......)))).))))....))......--- ( -26.40, z-score = -1.37, R) >droEre2.scaffold_4770 3255788 97 - 17746568 -UGCGUAUGUGUGCUUCUACACAUACAUAAGCUAAUACCAAC-ACAUACACACA--UGCAUGGAAAUGAACGUGGAAAAAUGUGAUUGUAGAAGGAGAGCG--- -.(((((((((((....))))))))).....((..(((...(-((((...(((.--..(((....)))...))).....)))))...)))..))....)).--- ( -25.30, z-score = -2.21, R) >droYak2.chr3R 11164205 97 + 28832112 -UGCGUCUGUGUGCUUCUGCACAUACAUAUGCUAAUACCAAC-ACACACACACU--CGCUUGAAAGUGGACGUGGAAAAAUGUGAGUGUAAAAGGAUAGCG--- -...((.(((((((....))))))).))..((((...((.((-((.((((((((--((((....)))))..)))......)))).))))....)).)))).--- ( -24.00, z-score = -0.28, R) >droSec1.super_0 6294222 99 - 21120651 -AGCGUCUGUGUGUUGCCACACACACAUAUGCUAAUACCAAC-ACACACACACAGACGCAUGGAAAUGGUCGUGGAAAAAUGUGAGUGUAAAAGGAGAGCG--- -(((((.(((((((......))))))).)))))....((.((-((.(((((((.(((.(((....)))))))))......)))).))))....))......--- ( -30.40, z-score = -1.73, R) >droSim1.chr3R 13286043 99 - 27517382 -AGCGUCUGUGUGUUGCCACACACACAUAUGCUAAUACCAAC-ACACACACACAGACGCAUGGAAAUGGUCGUGGAAAAAUGUGAGUGUAAAAGGAGAGCG--- -(((((.(((((((......))))))).)))))....((.((-((.(((((((.(((.(((....)))))))))......)))).))))....))......--- ( -30.40, z-score = -1.73, R) >droAna3.scaffold_13340 12999924 103 - 23697760 AUCAAUCAAUCAGAUUUAUUCGGCAAAAAGGUAGGUACUUUUUAUAUAUACAGUG-UCCUUGUUAAUAUCCUUAAGACAUUUCAAGUAUGAGGAGAAAGAAACC .((((((.....)))).....................((((((((((....((((-((.(((..........)))))))))....))))))))))...)).... ( -12.50, z-score = 1.05, R) >consensus _AGCGUCUGUGUGUUUCCACACACACAUAUGCUAAUACCAAC_ACACACACACA__CGCAUGGAAAUGGACGUGGAAAAAUGUGAGUGUAAAAGGAGAGCG___ .......(((((((....))))))).....(((....((....((((.((((......((((........))))......)))).))))....))..))).... (-14.87 = -14.85 + -0.02)
| Location | 7,128,322 – 7,128,419 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 72.15 |
| Shannon entropy | 0.51535 |
| G+C content | 0.42144 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -13.34 |
| Energy contribution | -12.68 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
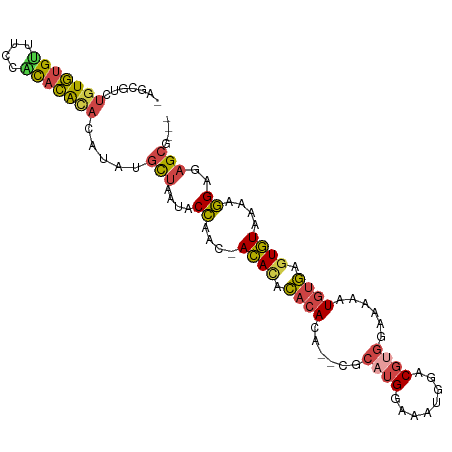
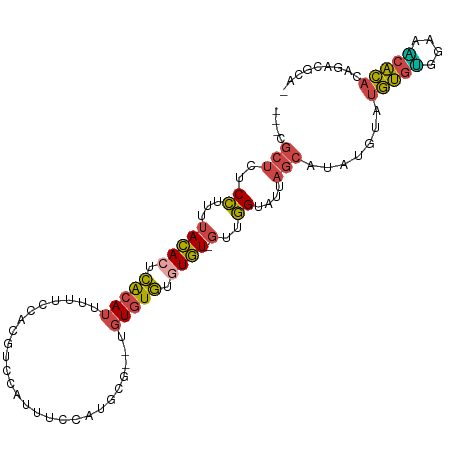
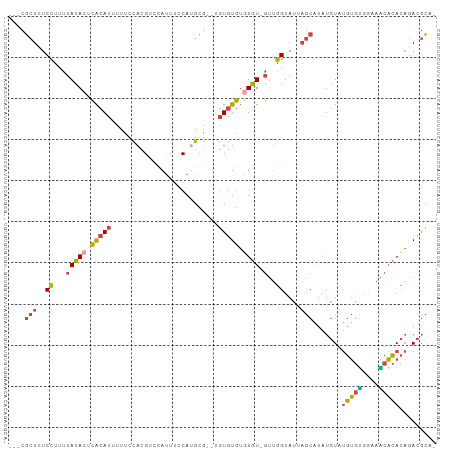
>dm3.chr3R 7128322 97 - 27905053 ---CGCUCUCCUUUUACACUCACAUUUUUCCACGUCCAUUUCCAUGCG--UGUGUGUGUGU-GUUGGUAUUAGCAUAUGUAUGUGUGGAAACACACAGACGCU- ---.(((..((....((((.(((((.....(((((..........)))--)).))))).))-)).))....)))....((.((((((....)))))).))...- ( -31.20, z-score = -2.91, R) >droEre2.scaffold_4770 3255788 97 + 17746568 ---CGCUCUCCUUCUACAAUCACAUUUUUCCACGUUCAUUUCCAUGCA--UGUGUGUAUGU-GUUGGUAUUAGCUUAUGUAUGUGUAGAAGCACACAUACGCA- ---.((....((..(((...(((((....((((((.(((....))).)--)))).).))))-)...)))..)).....((((((((......)))))))))).- ( -24.70, z-score = -2.15, R) >droYak2.chr3R 11164205 97 - 28832112 ---CGCUAUCCUUUUACACUCACAUUUUUCCACGUCCACUUUCAAGCG--AGUGUGUGUGU-GUUGGUAUUAGCAUAUGUAUGUGCAGAAGCACACAGACGCA- ---.((((.((...(((((.(((((((....................)--)))))).))))-)..))...))))....((.(((((....)))))...))...- ( -25.45, z-score = -1.00, R) >droSec1.super_0 6294222 99 + 21120651 ---CGCUCUCCUUUUACACUCACAUUUUUCCACGACCAUUUCCAUGCGUCUGUGUGUGUGU-GUUGGUAUUAGCAUAUGUGUGUGUGGCAACACACAGACGCU- ---.(((..((...(((((.(((((........((((((....))).))).))))).))))-)..))....)))....((.((((((....)))))).))...- ( -32.50, z-score = -2.55, R) >droSim1.chr3R 13286043 99 + 27517382 ---CGCUCUCCUUUUACACUCACAUUUUUCCACGACCAUUUCCAUGCGUCUGUGUGUGUGU-GUUGGUAUUAGCAUAUGUGUGUGUGGCAACACACAGACGCU- ---.(((..((...(((((.(((((........((((((....))).))).))))).))))-)..))....)))....((.((((((....)))))).))...- ( -32.50, z-score = -2.55, R) >droAna3.scaffold_13340 12999924 103 + 23697760 GGUUUCUUUCUCCUCAUACUUGAAAUGUCUUAAGGAUAUUAACAAGGA-CACUGUAUAUAUAAAAAGUACCUACCUUUUUGCCGAAUAAAUCUGAUUGAUUGAU (((((..(((........((((.((((((.....))))))..))))..-...........(((((((.......)))))))..))).)))))............ ( -12.10, z-score = 0.90, R) >consensus ___CGCUCUCCUUUUACACUCACAUUUUUCCACGUCCAUUUCCAUGCG__UGUGUGUGUGU_GUUGGUAUUAGCAUAUGUAUGUGUGGAAACACACAGACGCA_ ....(((..((..(.((((.(((((.....((.(........).)).....))))).)))).)..))....))).......(((((....)))))......... (-13.34 = -12.68 + -0.66)

| Location | 7,128,353 – 7,128,452 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 70.35 |
| Shannon entropy | 0.54806 |
| G+C content | 0.39464 |
| Mean single sequence MFE | -19.24 |
| Consensus MFE | -8.78 |
| Energy contribution | -8.79 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
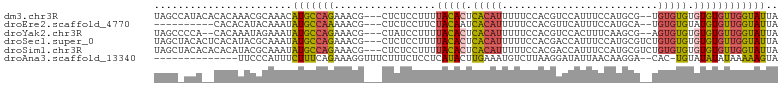
>dm3.chr3R 7128353 99 - 27905053 UAGCCAUACACACAAACGCAAACAUGCCAGAAACG---CUCUCCUUUUACACUCACAUUUUUCCACGUCCAUUUCCAUGCG--UGUGUGUGUGUGUUGGUAUUA ..((((.(((((((.(((((..((((......(((---...........................))).......))))..--))))).))))))))))).... ( -23.75, z-score = -1.93, R) >droEre2.scaffold_4770 3255819 89 + 17746568 ----------CACACAUACAAAUAUGCCAAAAACG---CUCUCCUUCUACAAUCACAUUUUUCCACGUUCAUUUCCAUGCA--UGUGUGUAUGUGUUGGUAUUA ----------.(((((((((.((((((....((((---...........................)))).........)))--))).)))))))))........ ( -15.55, z-score = -1.53, R) >droYak2.chr3R 11164236 97 - 28832112 UAGCCCCA--CACAAAUAGAAAUAUGCCAGAAACG---CUAUCCUUUUACACUCACAUUUUUCCACGUCCACUUUCAAGCG--AGUGUGUGUGUGUUGGUAUUA ..(((.((--((((.(((....)))..........---.........(((((((.(......................).)--))))))))))))..))).... ( -17.95, z-score = -0.77, R) >droSec1.super_0 6294253 101 + 21120651 UAGCUACACUCACAUACGCAAAUAUGCCAGAAACG---CUCUCCUUUUACACUCACAUUUUUCCACGACCAUUUCCAUGCGUCUGUGUGUGUGUGUUGGUAUUA ..(((((((.((((((((((....)))........---............................((((((....))).))).))))))).))).)))).... ( -21.80, z-score = -1.58, R) >droSim1.chr3R 13286074 101 + 27517382 UAGCUACACACACAUACGCAAAUAUGCCAGAAACG---CUCUCCUUUUACACUCACAUUUUUCCACGACCAUUUCCAUGCGUCUGUGUGUGUGUGUUGGUAUUA ..((((((((((((((((((....)))........---............................((((((....))).))).))))))))))).)))).... ( -25.80, z-score = -2.71, R) >droAna3.scaffold_13340 12999959 87 + 23697760 --------------UUCCCAUUUCUUUCAGAAAGGUUUCUUUCUCCUCAUACUUGAAAUGUCUUAAGGAUAUUAACAAGGA--CAC-UGUAUAUAUAAAAAGUA --------------..............((((((....)))))).......((((.((((((.....))))))..))))..--...-................. ( -10.60, z-score = 0.78, R) >consensus UAGC_ACA__CACAUACGCAAAUAUGCCAGAAACG___CUCUCCUUUUACACUCACAUUUUUCCACGUCCAUUUCCAUGCG__UGUGUGUGUGUGUUGGUAUUA .......................(((((((.................(((((.(((((.....((.(........).)).....))))).)))))))))))).. ( -8.78 = -8.79 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:52 2011