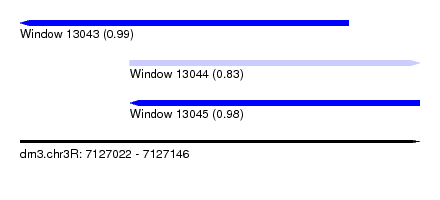
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,127,022 – 7,127,146 |
| Length | 124 |
| Max. P | 0.992809 |

| Location | 7,127,022 – 7,127,124 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.93 |
| Shannon entropy | 0.45812 |
| G+C content | 0.51914 |
| Mean single sequence MFE | -27.81 |
| Consensus MFE | -13.95 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
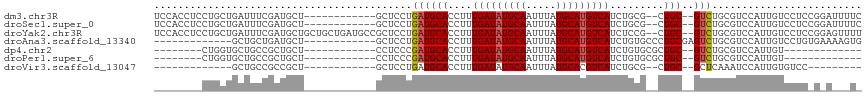
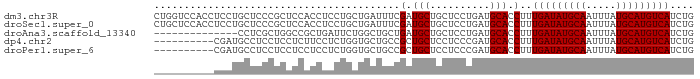
>dm3.chr3R 7127022 102 - 27905053 UCCACCUCCUGCUGAUUUCGAUGCU------------GCUCCUGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUGCG--CUGC--GUCUGCGUCCAUUGUCCUCCGGAUUUUC ......(((.(..(((...(((((.------------......((((((((..(((((((((.....)))))))))...).)--.)))--))).)))))....)))..).)))..... ( -27.40, z-score = -2.83, R) >droSec1.super_0 6292933 102 + 21120651 UCCACCUCCUGCUGAUUUCGAUGCU------------GCUCCUGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUGCG--CUGC--GUCUGCGUCCAUUGUCCUCCGGAUUUUC ......(((.(..(((...(((((.------------......((((((((..(((((((((.....)))))))))...).)--.)))--))).)))))....)))..).)))..... ( -27.40, z-score = -2.83, R) >droYak2.chr3R 11162844 114 - 28832112 UCCACCUCCUGCUGAUUUCGAUGCUGCUGCUGAUGCCGCUCCUGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUCCG--CUGC--GUCUGCGUCCAUUGUCCUCCGGAGUUUU .....((((.(..(((...(((((.((.((....)).))....((((((.(..(((((((((.....))))))))).....)--.)))--))).)))))....)))..).)))).... ( -34.80, z-score = -3.67, R) >droAna3.scaffold_13340 12998660 93 + 23697760 -------------GCUGCUGAUGCU------------GCUCCUGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUGUGCCCUGCGAGUCUGCGUCCAUUGUCCUGUGAAAAGUG -------------...((((((((.------------(((((....((((...(((((((((.....)))))))))...))))...).))))..)))))(((......)))...))). ( -29.00, z-score = -3.59, R) >dp4.chr2 1507967 83 + 30794189 --------CUGGUGCUGCCGCUGCU------------CCUCCCGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUGUGCGCUGC--GUCUGCGUCCAUUGU------------- --------.((((((.(.(((.(((------------(.....)).((((...(((((((((.....)))))))))...)))))).))--).).))).)))....------------- ( -29.60, z-score = -3.56, R) >droPer1.super_6 1529435 83 + 6141320 --------CUGGUGCUGCCGCUGCU------------CCUCCCGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUGUGCGCUGC--GUCUGCGUCCAUUGU------------- --------.((((((.(.(((.(((------------(.....)).((((...(((((((((.....)))))))))...)))))).))--).).))).)))....------------- ( -29.60, z-score = -3.56, R) >droVir3.scaffold_13047 15866635 80 + 19223366 -------------GCUGCCGCCGCU------------GCUCCUGAUGCACCUUUGAUAUGCAAUUUAUGCACGUCAUCUGCG--CUGC--GCUCAAAUCCAUUGUGUCC--------- -------------((.((.((.(((------------((.......)))....(((..((((.....))))..)))...)))--).))--)).................--------- ( -16.90, z-score = 0.13, R) >consensus ________CUGCUGCUGCCGAUGCU____________GCUCCUGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUGCG__CUGC__GUCUGCGUCCAUUGUCCUC_G_A___U_ ...........................................((((((....(((((((((.....)))))))))...((.....)).....))))))................... (-13.95 = -14.30 + 0.35)
| Location | 7,127,056 – 7,127,146 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 73.79 |
| Shannon entropy | 0.44292 |
| G+C content | 0.52902 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -12.24 |
| Energy contribution | -12.64 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
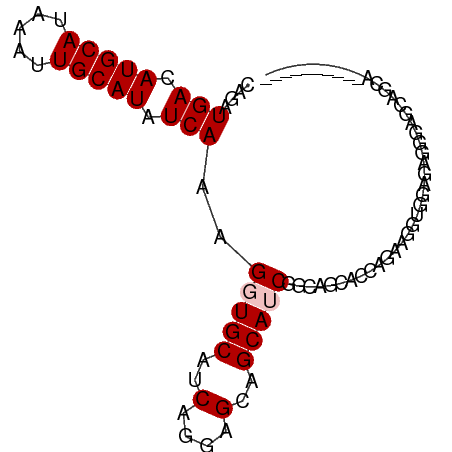
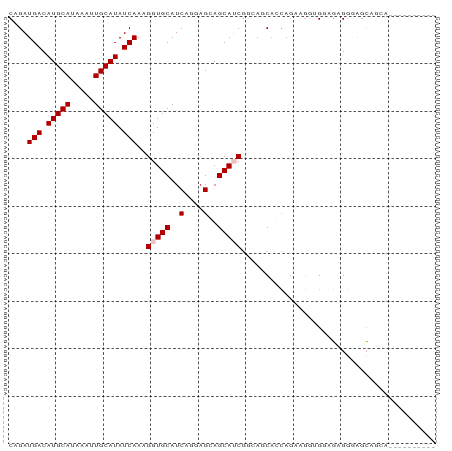
>dm3.chr3R 7127056 90 + 27905053 CAGAUGACAUGCAUAAAUUGCAUAUCAAAGGUGCAUCAGGAGCAGCAUCGAAAUCAGCAGGAGGUGGAGCGGGAGCAGGAGGUGGACCAG ....(((.(((((.....))))).)))..(((.((((........((((.............))))..((....))....)))).))).. ( -23.62, z-score = -2.07, R) >droSec1.super_0 6292967 90 - 21120651 CAGAUGACAUGCAUAAAUUGCAUAUCAAAGGUGCAUCAGGAGCAGCAUCGAAAUCAGCAGGAGGUGGAGCGGGAGCAGGAGGUGGAGCAG ....(((.(((((.....))))).)))..(.(.((((........((((.............))))..((....))....)))).).).. ( -19.12, z-score = -0.67, R) >droAna3.scaffold_13340 12998698 76 - 23697760 CAGAUGACAUGCAUAAAUUGCAUAUCAAAGGUGCAUCAGGAGCAGCAUCAGCAGCCAGAAUCAGCGGCCAGCGAGG-------------- ....(((.(((((.....))))).)))..(((((..........))))).((.(((.........)))..))....-------------- ( -17.90, z-score = -0.40, R) >dp4.chr2 1507990 80 - 30794189 CAGAUGACAUGCAUAAAUUGCAUAUCAAAGGUGCAUCGGGAGGAGCAGCGGCAGCACCAGAGGAAGAGGAGGAGGCAUCG---------- ..((((.((((((.....)))))......(((((.((....)).((....)).)))))...............).)))).---------- ( -20.60, z-score = -2.35, R) >droPer1.super_6 1529458 80 - 6141320 CAGAUGACAUGCAUAAAUUGCAUAUCAAAGGUGCAUCGGGAGGAGCAGCGGCAGCACCAGAGGAGGAGGAGGAGGCAUCG---------- ..((((.((((((.....)))))......(((((.((....)).((....)).)))))...............).)))).---------- ( -20.60, z-score = -2.31, R) >consensus CAGAUGACAUGCAUAAAUUGCAUAUCAAAGGUGCAUCAGGAGCAGCAUCGGCAGCACCAGAAGGUGGAGAGGGAGCAGCA__________ ....(((.(((((.....))))).)))..(((((..(....)..)))))......................................... (-12.24 = -12.64 + 0.40)

| Location | 7,127,056 – 7,127,146 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 73.79 |
| Shannon entropy | 0.44292 |
| G+C content | 0.52902 |
| Mean single sequence MFE | -17.34 |
| Consensus MFE | -11.94 |
| Energy contribution | -11.94 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
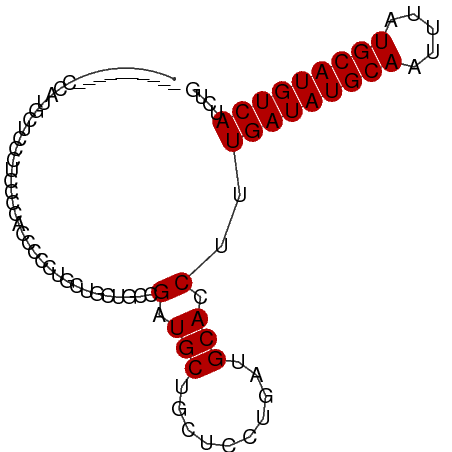
>dm3.chr3R 7127056 90 - 27905053 CUGGUCCACCUCCUGCUCCCGCUCCACCUCCUGCUGAUUUCGAUGCUGCUCCUGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUG ..(((.((.(....((....((((....((.....))....)).)).))....).)).)))..(((((((((.....))))))))).... ( -15.80, z-score = -1.14, R) >droSec1.super_0 6292967 90 + 21120651 CUGCUCCACCUCCUGCUCCCGCUCCACCUCCUGCUGAUUUCGAUGCUGCUCCUGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUG .(((..((......((((..((..........)).(....))).))......))..)))....(((((((((.....))))))))).... ( -14.50, z-score = -1.47, R) >droAna3.scaffold_13340 12998698 76 + 23697760 --------------CCUCGCUGGCCGCUGAUUCUGGCUGCUGAUGCUGCUCCUGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUG --------------....((.(((.(((......))).)))...))(((.......)))....(((((((((.....))))))))).... ( -20.40, z-score = -1.45, R) >dp4.chr2 1507990 80 + 30794189 ----------CGAUGCCUCCUCCUCUUCCUCUGGUGCUGCCGCUGCUCCUCCCGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUG ----------.(((..................(((((...((..........))..)))))...((((((((.....))))))))))).. ( -18.00, z-score = -2.85, R) >droPer1.super_6 1529458 80 + 6141320 ----------CGAUGCCUCCUCCUCCUCCUCUGGUGCUGCCGCUGCUCCUCCCGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUG ----------.(((..................(((((...((..........))..)))))...((((((((.....))))))))))).. ( -18.00, z-score = -2.93, R) >consensus __________CCAUGCUCCCUCCCCACCCCCUGCUGCUGCCGAUGCUGCUCCUGAUGCACCUUUGAUAUGCAAUUUAUGCAUGUCAUCUG .........................................(.(((..........))).)..(((((((((.....))))))))).... (-11.94 = -11.94 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:49 2011