
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,113,329 – 7,113,432 |
| Length | 103 |
| Max. P | 0.800320 |

| Location | 7,113,329 – 7,113,432 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 124 |
| Reading direction | forward |
| Mean pairwise identity | 64.21 |
| Shannon entropy | 0.68025 |
| G+C content | 0.45341 |
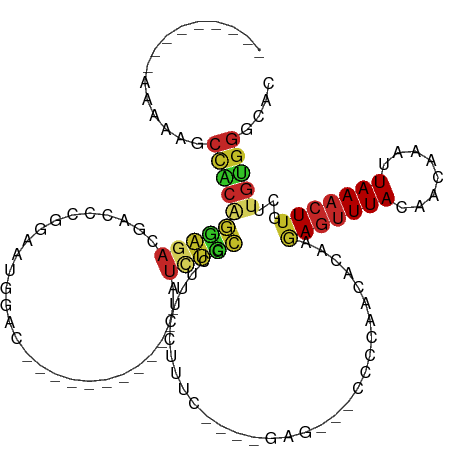
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -9.60 |
| Energy contribution | -9.08 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.800320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
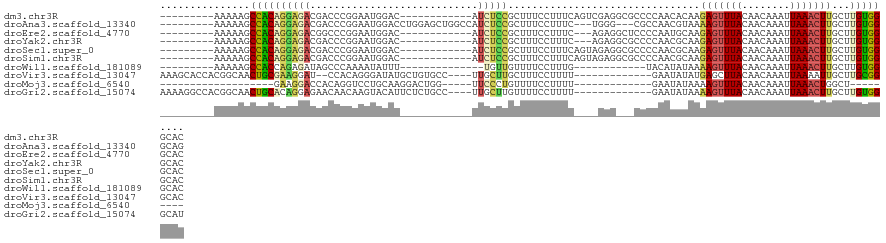
>dm3.chr3R 7113329 103 + 27905053 ---------AAAAAGCCACAGGAGACGACCCGGAAUGGAC------------AUCUCCGCUUUCCUUUCAGUCGAGGCGCCCCAACACAAGAGUUUACAACAAAUUAAACUUGCUUGUGGGCAC ---------.....(((...(....)(((..((((((((.------------...))))..)))).....)))..)))((((..(((...(((((((........)))))))...))))))).. ( -29.00, z-score = -1.96, R) >droAna3.scaffold_13340 12984313 109 - 23697760 ---------AAAAAGCCACAGGAGACGACCCGGAAUGGACCUGGAGCUGGCCAUCUCCGCUUUCCUUUC---UGGG---CGCCAACGUAAAAGUUUACAACAAAUUAAACUUGCUUGUGGGCAG ---------.....(((.((((((..((..((((((((.((.......)))))).))))..))..))))---))))---)(((.(((...(((((((........)))))))...))).))).. ( -36.00, z-score = -2.10, R) >droEre2.scaffold_4770 3240912 100 - 17746568 ---------AAAAAGCCACAGGAGACGGCCCGGAAUGGAC------------AUCUCCGCUUUCCUUUC---AGAGGCUCCCCAAUGCAAGAGUUUACAACAAAUUAAACUUGCUUGUGGGCAC ---------....((((...(((((.(.((......)).)------------.)))))....((.....---.)))))).((((..(((..((((((........)))))))))...))))... ( -25.70, z-score = -0.42, R) >droYak2.chr3R 11148784 100 + 28832112 ---------AAAAAGCCACAGGAGACGACCCGGAAUGGAC------------AUCUCCGCUUUCCUUUC---AGAGGCGCCCCAACGCAAGAGUUUACAACAAAUUAAACUUGCUUGUGGGCAC ---------.....(((...(....).....((((((((.------------...))))..))))....---...)))(((((((.(((..((((((........)))))))))))).)))).. ( -30.10, z-score = -2.36, R) >droSec1.super_0 6279406 103 - 21120651 ---------AAAAAGCCACAGGAGACGACCCGGAAUGGAC------------AUCUCCGCUUUCCUUUCAGUAGAGGCGCCCCAACGCAAGAGUUUACAACAAAUUAAACUUGCUUGUGGGCAC ---------.....(((...(((((.(.((......)).)------------.)))))(((........)))...)))(((((((.(((..((((((........)))))))))))).)))).. ( -30.90, z-score = -2.31, R) >droSim1.chr3R 13270494 103 - 27517382 ---------AAAAAGCCACAGGAGACGACCCGGAAUGGAC------------AUCUCCGCUUUCCUUUCAGUAGAGGCGCCCCAACGCAAGAGUUUACAACAAAUUAAACUUGCUUGUGGGCAC ---------.....(((...(((((.(.((......)).)------------.)))))(((........)))...)))(((((((.(((..((((((........)))))))))))).)))).. ( -30.90, z-score = -2.31, R) >droWil1.scaffold_181089 7999782 89 - 12369635 ---------AAAAAGCCACCAGAGAUAGCCCAAAAUAUUU--------------UGUUGUUUUCCUUUG------------UACAUAUAAAAGUUUACAACAAAUUAAACUUGCUUGUGGGCAC ---------.....(((..(((((..(((.((((....))--------------))..)))...)))))------------..((((...(((((((........)))))))...))))))).. ( -17.50, z-score = -1.37, R) >droVir3.scaffold_13047 15849619 105 - 19223366 AAAGCACCACGGCAACUGCGAAGGAU--CCACAGGGAUAUGCUGUGCC----UUGCUUGCUUUCCUUUU-------------GAAUAUAUGAGCUUACAACAAAUUAAAAUUGCUUGCGGGCAC ((((((.((.((((...(((....((--((....)))).)))..))))----.))..))))))......-------------..........(((..((((((.......))).)))..))).. ( -25.50, z-score = -0.30, R) >droMoj3.scaffold_6540 13752019 78 - 34148556 -------------------GAAGGACCACAGGUCCUGCAAGGACUGG-----UUCCCUGUUUUCCUUUU-------------GAAUAUAAAAGUUUACAACAAAUUAAACUGGCU--------- -------------------((((((..((((((((.....)))).(.-----...)))))..)))))).-------------.........((((((........))))))....--------- ( -16.30, z-score = -0.26, R) >droGri2.scaffold_15074 11897 107 - 7742996 AAAAGGCCACGGCAACUGCACAGGAGAACAACAAGUACAUUCUCUGCC----UUGCUUGUUUUCCUUUU-------------GAAUAUAAAAGUUUACAACAAAUUAAACUUGCUUGUGGGCAU ....(.(((((((........((((((((((((((............)----))).))))))))))...-------------........(((((((........))))))))).))))).).. ( -28.70, z-score = -2.43, R) >consensus _________AAAAAGCCACAGGAGACGACCCGGAAUGGAC____________AUCUCCGCUUUCCUUUC____GAG___CCCCAACACAAGAGUUUACAACAAAUUAAACUUGCUUGUGGGCAC ...............((((((((((............................)))))................................(((((((........)))))))...))))).... ( -9.60 = -9.08 + -0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:47 2011