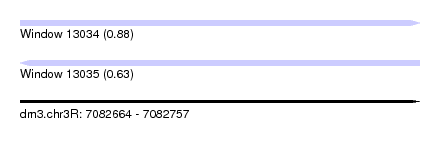
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,082,664 – 7,082,757 |
| Length | 93 |
| Max. P | 0.878458 |

| Location | 7,082,664 – 7,082,757 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Shannon entropy | 0.40563 |
| G+C content | 0.43737 |
| Mean single sequence MFE | -17.68 |
| Consensus MFE | -15.90 |
| Energy contribution | -15.52 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
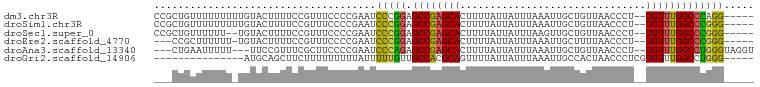
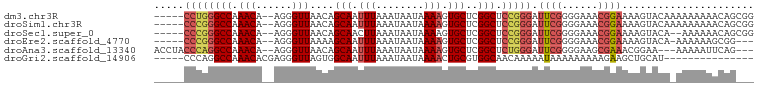
>dm3.chr3R 7082664 93 + 27905053 CCGCUGUUUUUUUUUGUACUUUUCCGUUUCCCCGAAUCCCGGAGCCGAGCACUUUUAUUAUUUAAAUUGCUGUUAACCCU--UGUUUGGCCCAGG----- ...(((................((((.(((...)))...))))((((((((.............................--)))))))).))).----- ( -13.65, z-score = 0.16, R) >droSim1.chr3R 13235460 93 - 27517382 CCGCUGUUUUUUUUUGUACUUUUCCGUUUCCCCGAAUCCCGGAGCCGAGCACUUUUAUUAUUUAAAUUGCUGUUAACCCU--UGUUUGGCCCGGG----- .....................................(((((.((((((((.............................--)))))))))))))----- ( -19.05, z-score = -1.27, R) >droSec1.super_0 6248553 91 - 21120651 CCGCUGUUUUUU--UGUACUUUUCCGUUUCCCCGAAUCCCGGAGCCGAGCACUUUUAUUAUUUAAGUUGCUGUUAACCCU--UGUUUGGCCCGGG----- ............--.......................(((((.((((((((...(((.....)))((((....))))...--)))))))))))))----- ( -20.00, z-score = -1.41, R) >droEre2.scaffold_4770 3208930 89 - 17746568 ---CCGCUUUUUU-UGUACUUUUCCGUUUCCCCGAAUCCCGGAGCCGAGCACUUUUAUUAUUUAAAUUGCUUUUAACCCU--UGUUUGGCCCGGG----- ---..........-.......................(((((.((((((((.............................--)))))))))))))----- ( -19.05, z-score = -1.73, R) >droAna3.scaffold_13340 12948118 92 - 23697760 ---CUGAAUUUUU---UUCCGUUUCGCUUCCCCGAAUCCCAGAGCCGAGCACUUUUAUUAUUUAAAUUGCUGUUAACCCU--UGUUUGGCCUGGGUAGGU ---..........---..((..((((......)))).(((((.((((((((.............................--)))))))))))))..)). ( -20.75, z-score = -1.21, R) >droGri2.scaffold_14906 14093693 80 - 14172833 ---------------AUGCAGCUUCUUUUUUUUUAUUUUUGUUGCCACGCAGUUUUAUUAUUUAAAUUGCCACUAACCCUCGUGUUUGGCCUGGG----- ---------------..(((((..................)))))...(((((((........)))))))......(((..((.....))..)))----- ( -13.57, z-score = -0.56, R) >consensus ___CUGUUUUUUU_UGUACUUUUCCGUUUCCCCGAAUCCCGGAGCCGAGCACUUUUAUUAUUUAAAUUGCUGUUAACCCU__UGUUUGGCCCGGG_____ .....................................(((((.((((((((......(((.............)))......)))))))))))))..... (-15.90 = -15.52 + -0.38)
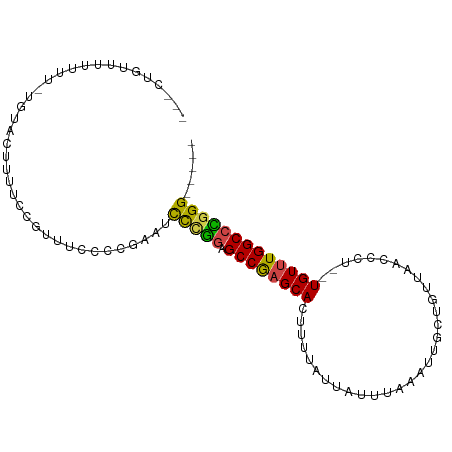
| Location | 7,082,664 – 7,082,757 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 77.64 |
| Shannon entropy | 0.40563 |
| G+C content | 0.43737 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -15.27 |
| Energy contribution | -16.22 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
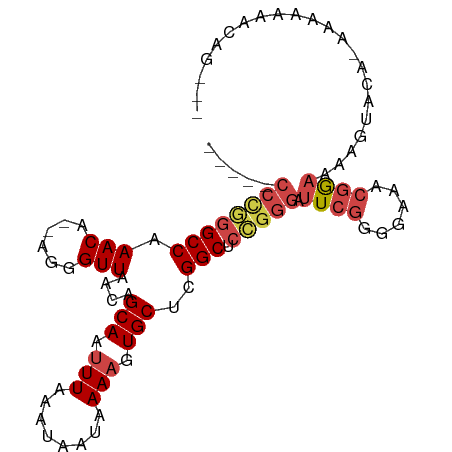
>dm3.chr3R 7082664 93 - 27905053 -----CCUGGGCCAAACA--AGGGUUAACAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACAAAAAAAAACAGCGG -----((((((((.(((.--...)))...((((.(((........))).)))).))).))))).((((......))))...................... ( -19.50, z-score = -0.84, R) >droSim1.chr3R 13235460 93 + 27517382 -----CCCGGGCCAAACA--AGGGUUAACAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACAAAAAAAAACAGCGG -----((((((((.(((.--...)))...((((.(((........))).)))).))).))))).((((......))))...................... ( -21.70, z-score = -1.27, R) >droSec1.super_0 6248553 91 + 21120651 -----CCCGGGCCAAACA--AGGGUUAACAGCAACUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACA--AAAAAACAGCGG -----((((((((.(((.--...)))...((((.(((.........))))))).))).))))).((((......))))........--............ ( -22.20, z-score = -1.39, R) >droEre2.scaffold_4770 3208930 89 + 17746568 -----CCCGGGCCAAACA--AGGGUUAAAAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACA-AAAAAAGCGG--- -----((((((((.(((.--...)))...((((.(((........))).)))).))).))))).((((......))))........-..........--- ( -21.70, z-score = -1.32, R) >droAna3.scaffold_13340 12948118 92 + 23697760 ACCUACCCAGGCCAAACA--AGGGUUAACAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCUGGGAUUCGGGGAAGCGAAACGGAA---AAAAAUUCAG--- .((..((((((((.(((.--...)))...((((.(((........))).)))).))).))))).((((......))))..))..---..........--- ( -22.00, z-score = -1.33, R) >droGri2.scaffold_14906 14093693 80 + 14172833 -----CCCAGGCCAAACACGAGGGUUAGUGGCAAUUUAAAUAAUAAAACUGCGUGGCAACAAAAAUAAAAAAAAAGAAGCUGCAU--------------- -----.....((((..(((........)))(((.(((........))).))).))))............................--------------- ( -11.30, z-score = 0.62, R) >consensus _____CCCGGGCCAAACA__AGGGUUAACAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACA_AAAAAAACAG___ .....((((((((.(((......)))....(((.(((........))).)))..))).))))).((((......))))...................... (-15.27 = -16.22 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:41 2011