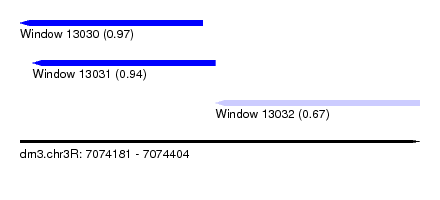
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,074,181 – 7,074,404 |
| Length | 223 |
| Max. P | 0.965215 |

| Location | 7,074,181 – 7,074,283 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.43 |
| Shannon entropy | 0.68193 |
| G+C content | 0.47054 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -12.04 |
| Energy contribution | -11.23 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7074181 102 - 27905053 -AUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUA-------AGCCAAGUGCAGUGCCGACUGACCCC--GCCCACAACAACGCCCCCCGUUGAGCAAAAAAAAAA----- -.............(((((.........(.(..(((((((.-------.....)))))))..))..........--........(((((.....))))))))))........----- ( -21.30, z-score = -2.08, R) >droSim1.chr3R 13226930 102 + 27517382 -AUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUU-------AGCCAAGUGCAGUGCCAACUGACCCC--GCCCACAACAACGCCCCCCGUUGAGCAAAAAAAAAA----- -.............(((((.........(.(..((((((((-------....))))))))..))..........--........(((((.....))))))))))........----- ( -21.80, z-score = -2.16, R) >droSec1.super_0 6240048 102 + 21120651 -AUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUU-------AGCCAAGUGCAGUGCCAACUGACCCC--GCCCACAACAACGCCCCCCGUUGAGCAAAAAAAAAA----- -.............(((((.........(.(..((((((((-------....))))))))..))..........--........(((((.....))))))))))........----- ( -21.80, z-score = -2.16, R) >droYak2.chr3R 11107530 100 - 28832112 -AUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUC-------AACCGAGUGCAGUGCCGACUGACCCC--GCCCAAAACAACGCUUCCAGCUGACCCCAAAAAC------- -.............(((.((((.((...(((((((((((((-------....)))))((((....)))).....--........))))))))..))))))...)))....------- ( -19.50, z-score = -1.25, R) >droEre2.scaffold_4770 3200574 93 + 17746568 -AUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUU-------AACCAAGUGCAGUGCCAAUUGACCCC--GCCCACAACAACGCCUACAA--AAACAAC------------ -.......((((..(((((((....)))).(..((((((((-------....))))))))..)...........--.....)))..))))......--.......------------ ( -16.30, z-score = -0.84, R) >droAna3.scaffold_13340 12939919 105 + 23697760 GAUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUUGC-GGGCACACAAGUGCAGGACCGACUGACCCC--GCCCACAAC--GCCCUCGUCCUUCUCUACCCCAC------- (((....((((((((..((((....))))...)))))))).((-((((((....)))(((......)))..)))--)).......--......)))..............------- ( -22.40, z-score = 0.26, R) >dp4.chr2 1492326 116 + 30794189 -AUACCUAGUGUAAUUGCUUAGUCCUAAGAGUGCUGCACUUGCAAGGAACCCAAGUGCAGGACUGACUGACCCAUGACCCACCACAGACCCCCCCACUCCUCCCCCCCAAAAUGUAU -.......(((..((.(.((((((.....(((.(((((((((.........))))))))).))))))))).).))....)))................................... ( -22.70, z-score = -1.25, R) >droPer1.super_6 1474009 116 + 6141320 -AUACCUAGUGUAAUUGCUUAGUCCUAAGAGUGCUGCACUUGCAAGGAACCCAAGUGCAGGACUGACUGACCCAUGACCCACCACAGACCCCCACACUCCUCCCCCCCAAAAUGUAU -......(((((......((((((.....(((.(((((((((.........))))))))).)))))))))....((......)).........)))))................... ( -24.00, z-score = -1.41, R) >droWil1.scaffold_181089 7954868 106 + 12369635 -AUACCUAGUGUAAUUGCUUAGACUUAAGAGUUUUGCACUUGC-GGGAAGACAAGUGCAGGACCUGACCCCCCC--UCCCGGACCCACAGAUCUAAUUUCACACUCAAGC------- -......(((((......(((((((..((.(((((((((((((-.....).)))))))))))))).......((--....))......)).)))))....))))).....------- ( -24.30, z-score = -0.81, R) >droVir3.scaffold_13047 15804838 106 + 19223366 -AUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGUACUUGC-AGGAAGACAAGUGCAAGACCCAAUAACCAGGGCAAUAUA--GUUAGCCAAGAUUAGCUCAAAAGUA------- -....((((.((....))))))((((..((((.(((((((((.-.......)))))))))(.(((........))))......--..............))))..)))).------- ( -21.60, z-score = -0.04, R) >droMoj3.scaffold_6540 13705561 105 + 34148556 -AUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGUACUUGC-GGGAAGACAAGUGCAAGACCCAACAACCAGGGCCAGAUUAUA-UAGAAAUGCCUA--UAGAAAGUU------- -...((((((.(((....))).)))...(.((.((((((((((-.....).))))))))).)).).......))).........((-(((......)))--)).......------- ( -21.20, z-score = -0.34, R) >droGri2.scaffold_14906 14085974 107 + 14172833 -AUACCUAGUGUAAUUGCUUAGACUUGAGAGUGUUGUACUUGC-GGGAAGACAAGUGCAAGACCCAA-AACCAGGGUAACAUAGUGUUAGCCAAG-UUAGUUCAAAAGUAG------ -.(((....((.((((((((....(((.(.((.((((((((((-.....).))))))))).))))))-.....((.((((.....)))).)))))-.)))))))...))).------ ( -25.80, z-score = -0.94, R) >consensus _AUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUUGC__GGAAGCCAAGUGCAGGACCGACUGACCCC__GCCCACAACAACGCCCCCCAUUAAUCAAAAAAAA_______ .....((((.((....))))))........((.((((((((...........)))))))).))...................................................... (-12.04 = -11.23 + -0.81)
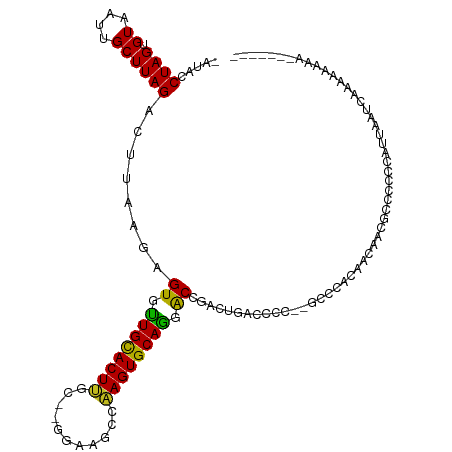
| Location | 7,074,188 – 7,074,290 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.75 |
| Shannon entropy | 0.64433 |
| G+C content | 0.50327 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -15.15 |
| Energy contribution | -14.31 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7074188 102 - 27905053 -GUUGGGGAUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUA-------AGCCAAGUGCAGUGCCGACUGACCC--CGCCCACAACAACGCCCCCCGUUGAGCAAA----- -(((((((.....(((((((((..((((....))))...)))))))))-------.........((((....)))).)))--)........(((((.....))))))))...----- ( -27.70, z-score = -1.26, R) >droSim1.chr3R 13226937 102 + 27517382 -GUUGGGGAUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUU-------AGCCAAGUGCAGUGCCAACUGACCC--CGCCCACAACAACGCCCCCCGUUGAGCAAA----- -(((((((......(((.(((....))).)))...(.(..((((((((-------....))))))))..))......)))--)........(((((.....))))))))...----- ( -28.10, z-score = -1.32, R) >droSec1.super_0 6240055 102 + 21120651 -GUUGGGGAUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUU-------AGCCAAGUGCAGUGCCAACUGACCC--CGCCCACAACAACGCCCCCCGUUGAGCAAA----- -(((((((......(((.(((....))).)))...(.(..((((((((-------....))))))))..))......)))--)........(((((.....))))))))...----- ( -28.10, z-score = -1.32, R) >droYak2.chr3R 11107535 102 - 28832112 -GUUGGGGAUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUC-------AACCGAGUGCAGUGCCGACUGACCC--CGCCCAAAACAACGCUUCCAGCUGACCCCA----- -..(((((.((.(((((((.....((((....)))).(..((((((((-------....))))))))..)..........--...........)))))...)).)).)))))----- ( -27.60, z-score = -1.33, R) >droEre2.scaffold_4770 3200579 95 + 17746568 -GUUGGGGAUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUU-------AACCAAGUGCAGUGCCAAUUGACCC--CGCCCACAACAACGCCUACAAAA------------ -(((((((.......((.((((((((((....)))).(..((((((((-------....))))))))..)))))))))..--..))).)))).............------------ ( -24.20, z-score = -1.22, R) >droAna3.scaffold_13340 12939919 112 + 23697760 GUUGGGGGAUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUUGC-GGGCACACAAGUGCAGGACCGACUGACCC--CGCCCACAACG--CCCUCGUCCUUCUCUACCCCAC ..(((((.......((((((((..((((....))))...)))))))).((-((((((....)))(((......)))..))--)))........--................))))). ( -31.30, z-score = 0.07, R) >dp4.chr2 1492333 115 + 30794189 --GUGGGGAUACCUAGUGUAAUUGCUUAGUCCUAAGAGUGCUGCACUUGCAAGGAACCCAAGUGCAGGACUGACUGACCCAUGACCCACCACAGACCCCCCCACUCCUCCCCCCCAA --((((((....((.(((..((.(.((((((.....(((.(((((((((.........))))))))).))))))))).).))....)))...))....))))))............. ( -33.50, z-score = -2.01, R) >droPer1.super_6 1474016 115 + 6141320 --GUGGGGAUACCUAGUGUAAUUGCUUAGUCCUAAGAGUGCUGCACUUGCAAGGAACCCAAGUGCAGGACUGACUGACCCAUGACCCACCACAGACCCCCACACUCCUCCCCCCCAA --((((((.......(((..((.(.((((((.....(((.(((((((((.........))))))))).))))))))).).))....))).......))))))............... ( -34.74, z-score = -2.21, R) >droWil1.scaffold_181089 7954873 108 + 12369635 -GUUGGGGAUACCUAGUGUAAUUGCUUAGACUUAAGAGUUUUGCACUUGC-GGGAAGACAAGUGCAGGACCUGACCCCCC--CUCCCGGACCCACAGAUCUAAUUUCACACU----- -(((.((((.....(((.(((....))).)))..((.(((((((((((((-.....).))))))))))))))........--.)))).))).....................----- ( -29.10, z-score = -0.68, R) >droVir3.scaffold_13047 15804844 107 + 19223366 -GUUGGGGAUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGUACUUGC-AGGAAGACAAGUGCAAGACCCAAUAACCAGGGCAAUAUA--GUUAGCCAAGAUUAGCUCA------ -((((((((((((((((.(((....))).)))..)).)))))(((((((.-.......)))))))....)))))).....((((....((--(((......))))))))).------ ( -26.80, z-score = -0.59, R) >droMoj3.scaffold_6540 13705567 106 + 34148556 -GUUGGGGAUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGUACUUGC-GGGAAGACAAGUGCAAGACCCAACAACCAGGGCCAGAUUAUA-UAGAAAUGCCUAUAG-------- -((((((((((((((((.(((....))).)))..)).)))))((((((((-.....).)))))))....)))))).....((((.........-.......))))....-------- ( -28.39, z-score = -1.42, R) >droGri2.scaffold_14906 14085981 107 + 14172833 -GUUGGGGAUACCUAGUGUAAUUGCUUAGACUUGAGAGUGUUGUACUUGC-GGGAAGACAAGUGCAAGACCCAA-AACCAGGGUAACAUAGUGUUAGCCAAG-UUAGUUCA------ -.(((((((((((((((.(((....))).)))..)).)))))((((((((-.....).)))))))....)))))-(((..((.((((.....)))).))..)-))......------ ( -29.20, z-score = -0.86, R) >consensus _GUUGGGGAUACCUAGUGUAAUUGCUUAGACUUAAGAGUGUUGCACUUGC__GGAAGCCAAGUGCAGGACCGACUGACCC__CGCCCACAACAACGCCCCCCAUUGAUCCA______ ...((((....))))(((......((((....)))).((.((((((((...........)))))))).))................)))............................ (-15.15 = -14.31 + -0.84)
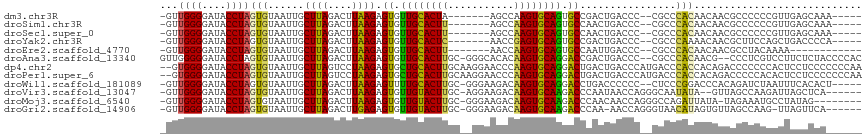
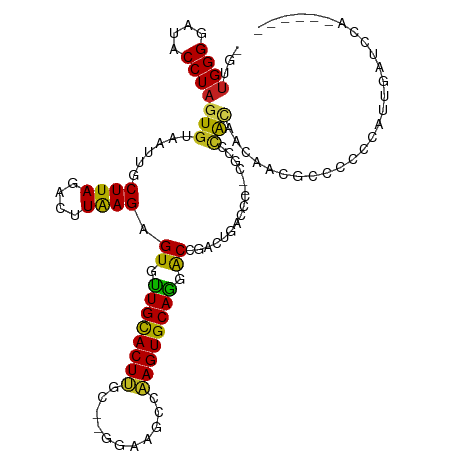
| Location | 7,074,290 – 7,074,404 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.68 |
| Shannon entropy | 0.16923 |
| G+C content | 0.42231 |
| Mean single sequence MFE | -19.95 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.13 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.668478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7074290 114 - 27905053 ACAGCCAAACAAAAGGACAGGUAGUAUGCGAGUAUAGCAUAUAUGCAUAUAUUCCAUAGCCAGAACAGGAGUAAAAGGACCACAAUCGCGAAAAGUUAACAUUUCGGACACAAG ...(((........)).).(((..((((.((((((((((....))).))))))))))).((......)).........)))....((.(((((.(....).)))))))...... ( -19.90, z-score = -1.84, R) >droYak2.chr3R 11107637 105 - 28832112 ACAGCCAAACGAUAGGACAGGAAAUGUGCGCAUACA--------GCAUAUAUUCCAUAGCCAGAACAGGAG-AUAAGGACCAAAAUCGCGAAAAGUAAACAUUUCGGACACAAG ...(((...((((.((...((((((((((.......--------)))))).))))((..((......))..-)).....))...)))).((((.(....).)))))).)..... ( -19.50, z-score = -2.42, R) >droSec1.super_0 6240157 112 + 21120651 ACAGCCAAACGAAAGGACAGGGAGUAUGCGAGUAUAGC--AUAUGCAUAUAUUCCAUAGCCAGAACAGGAGAAAAAGGACCAGAAUCGCGAAAAGAUAACAUUUCGGACACAAG ....((...(....).....((..((((.(((((((((--....)).))))))))))).))......))................((.(((((........)))))))...... ( -20.20, z-score = -2.23, R) >droSim1.chr3R 13227039 112 + 27517382 ACAGCCAAACGAAAGGACAGGGAGUAUGCGAGUAUAGC--AUAUGCAUAUAUUCCAUAGCCAGAACAGGAGAAAAAGGACCAGAAUCGCGAAAAGAUAACAUUUCGGACACAAG ....((...(....).....((..((((.(((((((((--....)).))))))))))).))......))................((.(((((........)))))))...... ( -20.20, z-score = -2.23, R) >consensus ACAGCCAAACGAAAGGACAGGGAGUAUGCGAGUAUAGC__AUAUGCAUAUAUUCCAUAGCCAGAACAGGAGAAAAAGGACCAGAAUCGCGAAAAGAUAACAUUUCGGACACAAG ..............((...((((((((((...............)))))).))))....((......))..........))....((.(((((.(....).)))))))...... (-14.39 = -14.13 + -0.25)

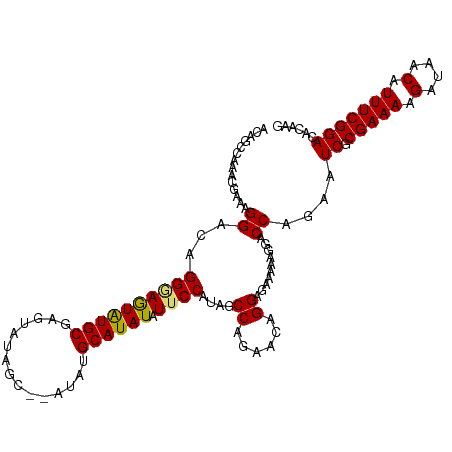
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:38 2011