
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,073,834 – 7,073,930 |
| Length | 96 |
| Max. P | 0.999732 |

| Location | 7,073,834 – 7,073,930 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.03 |
| Shannon entropy | 0.30724 |
| G+C content | 0.31278 |
| Mean single sequence MFE | -25.81 |
| Consensus MFE | -14.91 |
| Energy contribution | -14.67 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.998953 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
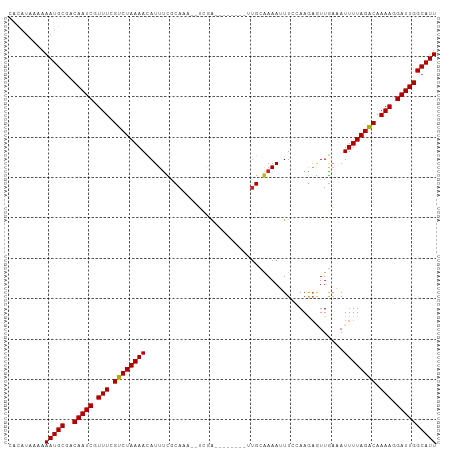
>dm3.chr3R 7073834 96 + 27905053 CACAUAAAAAAUGCGGCAAUCGUUUCGUCUAAAAUAUUCCGCAAU--UCAA--------UUGCGAAAUUUCCAAGAGUUGAAAUUUUAGACAAAAGGAUUGGCAUU .........(((((..(((((.(((.(((((((((.(((((((((--...)--------))))).(((((....))))))))))))))))).))).)))))))))) ( -25.50, z-score = -3.64, R) >droSim1.chr3R 13226591 96 - 27517382 CACAUAAAAAAUGCGGCAAUCGUUUCGUCUAAAACAUUUCGCAAU--UCGA--------UUGCAAAAUUUCCAAAAGUUGAAAUUUUAGACAAAAGGAUUGGCAUU .........(((((..(((((.(((.(((((((..((((((((((--...)--------))))..(((((....))))))))))))))))).))).)))))))))) ( -23.30, z-score = -3.15, R) >droSec1.super_0 6239706 96 - 21120651 CACAUAAAAAAUGCGGCAAUCGUUUCGUCUAAAACAUUUCGCAAU--UCGA--------UUGCGAAAUUUCCAAAAGUUGAAAUUUUAGACAAAAGGAUUGGCAUU .........(((((..(((((.(((.((((((((..(((((((((--...)--------))))))))((((........)))))))))))).))).)))))))))) ( -27.20, z-score = -4.07, R) >droYak2.chr3R 11107190 96 + 28832112 CACAUAAAAAAUGCGACAAUCGUUUCGUCUAAAACAUUUCGCAAU--UCGA--------UUGCGAAAUUUCCAAGUCUUGAAAUUUUAGGCAAAAGGAUUGGCAUU .........(((((..(((((.(((.((((((((..(((((((((--...)--------))))))))((((........)))))))))))).))).)))))))))) ( -26.60, z-score = -3.75, R) >droEre2.scaffold_4770 3200234 96 - 17746568 CACAUAAAAAAUGCGACAAUCGUUUCGUCUAAAACAUUUCACAAU--UCGA--------UUGCGAAAUUUCCAAGUGUUGAAAUUUUAGGCAAAAGGAUUGGCAUU .........(((((..(((((.(((.(((((((..((((((((..--....--------(((.((....))))).)).))))))))))))).))).)))))))))) ( -21.70, z-score = -2.15, R) >droAna3.scaffold_13340 12939587 96 - 23697760 UACAUAAAAAAUGCGACAAUCGUUUCGUCUAAAACAUUUCGCAAA--UAAA--------AAGCGAAAUUUCCAAGAAUUGAAAUUUUAGGCAAAAGGAUUGGCAUU .........(((((..(((((.(((.((((((((..((((((...--....--------..))))))((((........)))))))))))).))).)))))))))) ( -23.80, z-score = -4.00, R) >droWil1.scaffold_181089 7954446 106 - 12369635 CAUAUAAAAAAUGCGGCAAUCGUUUCGUCUAAAAUAUUUUGCAAAAGUCAAAGUUAUUUUUGCAAAAAUUAGAAGAAUUGAUAUUUUAGGCAAAAGGAUUGGCAUU .........(((((..(((((.(((.((((((((((((((((((((((.......)))))))))).((((.....)))))))))))))))).))).)))))))))) ( -29.00, z-score = -4.68, R) >droVir3.scaffold_13047 15804521 96 - 19223366 UAUAUAAAAAAUGCAACAAUCGUUUCGUCUAAAACAUUUUGCACAAUACUG---------UGGAAAA-UUAGAAGAAUUGUCAUUUUAGACAAAAGGAUUGGCAUU .........(((((..(((((.(((.((((((((.(((((.((((....))---------)).))))-).....((....)).)))))))).))).)))))))))) ( -24.10, z-score = -4.33, R) >droMoj3.scaffold_6540 13705221 96 - 34148556 UAUAUAAAAAAUGCGACAAUCGUUUCGUCUAAAACAUUUUCCACAAGACUG---------UGCAAAA-UUAGAAGAGUUAACAUUUUAGACAAAACGAUUGGCAUU .........(((((..(((((((((.((((((((.(((((.((((....))---------)).))))-).......(....).)))))))).)))))))))))))) ( -24.80, z-score = -4.82, R) >droGri2.scaffold_14906 14085648 96 - 14172833 UAUAUAAAAAAUGCGACAAUCCUUUCGUCUAAAACAUUUUGCACAAGAUCG---------UGCAAAA-UUAGAAGAAUUGUCAUUUUAGACAAAAGGAUUGGCAUU .........(((((..(((((((((.((((((((.(((((((((......)---------)))))))-).....((....)).)))))))).)))))))))))))) ( -32.10, z-score = -6.89, R) >consensus CACAUAAAAAAUGCGACAAUCGUUUCGUCUAAAACAUUUCGCAAA__UCGA________UUGCAAAAUUUCCAAGAGUUGAAAUUUUAGACAAAAGGAUUGGCAUU .........(((((..(((((.(((.((((((((.................................................)))))))).))).)))))))))) (-14.91 = -14.67 + -0.24)


| Location | 7,073,834 – 7,073,930 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Shannon entropy | 0.30724 |
| G+C content | 0.31278 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -17.77 |
| Energy contribution | -17.26 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.17 |
| Mean z-score | -5.09 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.27 |
| SVM RNA-class probability | 0.999732 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
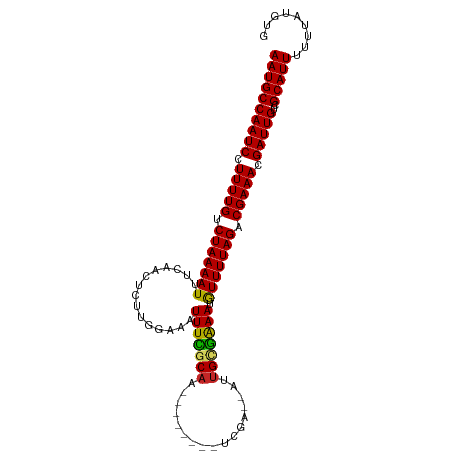
>dm3.chr3R 7073834 96 - 27905053 AAUGCCAAUCCUUUUGUCUAAAAUUUCAACUCUUGGAAAUUUCGCAA--------UUGA--AUUGCGGAAUAUUUUAGACGAAACGAUUGCCGCAUUUUUUAUGUG ((((((((((.(((((((((((((((((.....)))))(((((((((--------(...--)))))))))).)))))))))))).)))))..)))))......... ( -30.80, z-score = -5.57, R) >droSim1.chr3R 13226591 96 + 27517382 AAUGCCAAUCCUUUUGUCUAAAAUUUCAACUUUUGGAAAUUUUGCAA--------UCGA--AUUGCGAAAUGUUUUAGACGAAACGAUUGCCGCAUUUUUUAUGUG ((((((((((.(((((((((((((((((.....)))))(((((((((--------(...--)))))))))).)))))))))))).)))))..)))))......... ( -29.10, z-score = -4.92, R) >droSec1.super_0 6239706 96 + 21120651 AAUGCCAAUCCUUUUGUCUAAAAUUUCAACUUUUGGAAAUUUCGCAA--------UCGA--AUUGCGAAAUGUUUUAGACGAAACGAUUGCCGCAUUUUUUAUGUG ((((((((((.(((((((((((((((((.....)))))(((((((((--------(...--)))))))))).)))))))))))).)))))..)))))......... ( -31.20, z-score = -5.50, R) >droYak2.chr3R 11107190 96 - 28832112 AAUGCCAAUCCUUUUGCCUAAAAUUUCAAGACUUGGAAAUUUCGCAA--------UCGA--AUUGCGAAAUGUUUUAGACGAAACGAUUGUCGCAUUUUUUAUGUG ((((((((((.(((((.(((((((((((.....)))))(((((((((--------(...--)))))))))).)))))).))))).)))))..)))))......... ( -26.20, z-score = -3.43, R) >droEre2.scaffold_4770 3200234 96 + 17746568 AAUGCCAAUCCUUUUGCCUAAAAUUUCAACACUUGGAAAUUUCGCAA--------UCGA--AUUGUGAAAUGUUUUAGACGAAACGAUUGUCGCAUUUUUUAUGUG ((((((((((.(((((.(((((((((((.....)))))(((((((((--------(...--)))))))))).)))))).))))).)))))..)))))......... ( -24.30, z-score = -3.23, R) >droAna3.scaffold_13340 12939587 96 + 23697760 AAUGCCAAUCCUUUUGCCUAAAAUUUCAAUUCUUGGAAAUUUCGCUU--------UUUA--UUUGCGAAAUGUUUUAGACGAAACGAUUGUCGCAUUUUUUAUGUA ((((((((((.(((((.(((((((((((.....)))))(((((((..--------....--...))))))).)))))).))))).)))))..)))))......... ( -23.90, z-score = -3.68, R) >droWil1.scaffold_181089 7954446 106 + 12369635 AAUGCCAAUCCUUUUGCCUAAAAUAUCAAUUCUUCUAAUUUUUGCAAAAAUAACUUUGACUUUUGCAAAAUAUUUUAGACGAAACGAUUGCCGCAUUUUUUAUAUG ((((((((((.(((((.(((((((((..............(((((((((...........)))))))))))))))))).))))).)))))..)))))......... ( -23.13, z-score = -4.71, R) >droVir3.scaffold_13047 15804521 96 + 19223366 AAUGCCAAUCCUUUUGUCUAAAAUGACAAUUCUUCUAA-UUUUCCA---------CAGUAUUGUGCAAAAUGUUUUAGACGAAACGAUUGUUGCAUUUUUUAUAUA ((((((((((.(((((((((((((((....)).....(-((((.((---------(......))).)))))))))))))))))).)))))..)))))......... ( -24.20, z-score = -4.86, R) >droMoj3.scaffold_6540 13705221 96 + 34148556 AAUGCCAAUCGUUUUGUCUAAAAUGUUAACUCUUCUAA-UUUUGCA---------CAGUCUUGUGGAAAAUGUUUUAGACGAAACGAUUGUCGCAUUUUUUAUAUA ((((((((((((((((((((((((.............(-((((.((---------(......))).))))))))))))))))))))))))..)))))......... ( -31.31, z-score = -6.39, R) >droGri2.scaffold_14906 14085648 96 + 14172833 AAUGCCAAUCCUUUUGUCUAAAAUGACAAUUCUUCUAA-UUUUGCA---------CGAUCUUGUGCAAAAUGUUUUAGACGAAAGGAUUGUCGCAUUUUUUAUAUA ((((((((((((((((((((((((((....)).....(-(((((((---------((....)))))))))))))))))))))))))))))..)))))......... ( -37.00, z-score = -8.56, R) >consensus AAUGCCAAUCCUUUUGUCUAAAAUUUCAACUCUUGGAAAUUUCGCAA________UCGA__AUUGCGAAAUGUUUUAGACGAAACGAUUGUCGCAUUUUUUAUGUG ((((((((((.(((((.(((((((...............(((((((.................))))))).))))))).))))).)))))..)))))......... (-17.77 = -17.26 + -0.51)

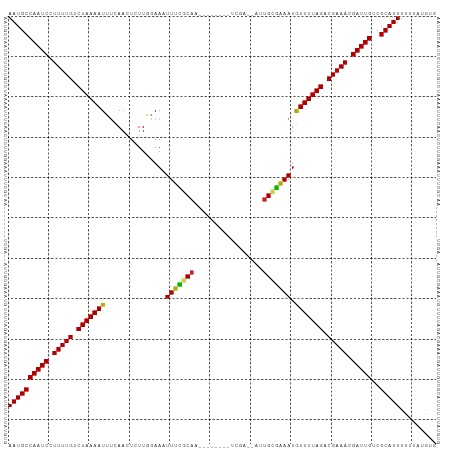
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:36 2011