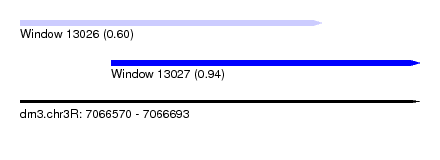
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,066,570 – 7,066,693 |
| Length | 123 |
| Max. P | 0.935660 |

| Location | 7,066,570 – 7,066,663 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.24 |
| Shannon entropy | 0.62165 |
| G+C content | 0.51664 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -17.67 |
| Energy contribution | -16.56 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.75 |
| Mean z-score | -0.38 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.601834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7066570 93 + 27905053 ----------CAGCUAUCACUA-CUUACUCCCUUGUUGUUGCAGGCUAUGUGCUUGUCCAGGCACGUGGGCAGAGCACAAUAGUCCGCUGCAAGGCUUAUUCCC--------- ----------............-.......((((((....((.(((((((((((((((((......)))))).))))).)))))).)).)))))).........--------- ( -32.00, z-score = -1.58, R) >droSim1.chr3R 14317669 93 + 27517382 ----------CAGCUACCACUA-CUUACUCCCUUGUUGUUGCAGGCUAUGUGCUUGUCCAGGCAAGUGGGCAGGGCACAGUAGUCUGCUGCAAGGCUCAAUCCC--------- ----------............-.......((((((....((((((((((((((((((((......)))))).)))))).)))))))).)))))).........--------- ( -36.30, z-score = -1.47, R) >droSec1.super_0 6228145 93 - 21120651 ----------CAGCUACCACUA-CUUACUCCCUUGUUGUUGCAGGCUAUGUGCUUGUCCAGGCAAGUGGGCAGGGCACAGUAGUCCGCUGCAAGGCUCAAUCCC--------- ----------............-.......((((((....((.(((((((((((((((((......)))))).)))))).))))).)).)))))).........--------- ( -31.70, z-score = -0.26, R) >droYak2.chr3R 11099646 93 + 28832112 ----------UACCUAUCACUA-CUUACUCCCUUAUUGUUGCAGGCAAUGUGCUUGUCCAGGCAAGUGGGCAGAGCACAGUAGUCCGCUGCAAGGCUCAAUCCC--------- ----------............-.......(((...(((.((.(((..((((((((((((......)))))).))))))...))).)).)))))).........--------- ( -25.80, z-score = -0.08, R) >droEre2.scaffold_4770 3192855 93 - 17746568 ----------UACCUAUCCCUA-CUUACUCCCUUAUUAUUGCAGGCAAUGUGCUUGUCCAGGCAAGUGGGCAGGGCACAGUAGUCCGCUGCGAGGCUCAAUCCC--------- ----------........((((-(((....(((.......((((((.....))))))..))).)))))))..((((...((((....))))...))))......--------- ( -25.60, z-score = 0.50, R) >droAna3.scaffold_13340 12930350 94 - 23697760 ---------UAGACCAAGAGUU-CCUACCCCCUUGUUGUUGCAGGCAAUGUGCUUGUCCAGGCAGGUGGGCAGAGCACAGUAGUCGACUGCGAAGGCAAUGCCC--------- ---------..(.((....(((-(...(((.((((((...((((((.....))))))...)))))).)))..))))...((((....))))...))).......--------- ( -29.50, z-score = -0.03, R) >dp4.chr2 2203094 92 - 30794189 -----------AGCGUCUGGCA-CUUACUCCUUUAUUAUUGCAGGCCACGUGCUUGUCCAUGCAGGUGGGCAGUGCACAGUAGUCGACGGCGAUGGCAGCUCCU--------- -----------((((((.((..-......))......(((((.(((.((((((((((((((....)))))))).)))).)).)))....)))))))).)))...--------- ( -28.10, z-score = 1.05, R) >droPer1.super_6 3916868 92 + 6141320 -----------AGCGUCUGGCA-CUUACUCCUUUAUUAUUGCAGGCCACGUGCUUGUCCAUGCAGGUGGGCAGUGCACAGUAGUCGACGGCGAUGGCAGCUCCU--------- -----------((((((.((..-......))......(((((.(((.((((((((((((((....)))))))).)))).)).)))....)))))))).)))...--------- ( -28.10, z-score = 1.05, R) >droWil1.scaffold_181089 2348038 107 + 12369635 UUGUUAAUAUUUUAAUGAAAAAACUUACUCCUUGAGAAGUACAGGCCAAAUUUAAGCCUUGGCAGUGGGGAUUCUGACAAUAGUUUGGAAUUCUUGCCAUUGAAUUU------ ..........(((((((.....((((.(((...)))))))....(((((.........))))).(..(((((((..(.......)..)))))))..))))))))...------ ( -24.60, z-score = -1.19, R) >droVir3.scaffold_13047 391496 103 - 19223366 ----------UAUCCUUAACCAACUUACUCCCUUGUUGUUGCAGGCAAUGUGCUUGUCCAAGCACGUGGGCAGAGCACAGUAGUCCACUGCAUUGGCAAUUUUUGCCAGACCA ----------...(((((((.(((..........))))))).)))...((((((((((((......)))))).))))))((((....)))).((((((.....)))))).... ( -31.00, z-score = -1.17, R) >droMoj3.scaffold_6540 2988991 93 + 34148556 ----------AAGGAUUAAAUA-CUUACUCCUUUAUUAUUGCAGGCAAUGUGCUUGUCCAGGCAGGUGGGCAAGGCGCAGUAGUCCACGGAGGCAACGAGGCCU--------- ----------..((((((....-......(((.((....)).)))...((((((((((((......))))))).))))).))))))....((((......))))--------- ( -27.70, z-score = -0.27, R) >droGri2.scaffold_14830 3294237 100 - 6267026 ----ACUAUUAGACUAUAAGUCACUUACUCCCUUGUUGUUGCAGGCAAUGUGCUUGUCCAGGCAGGUGGGCAAGGCACAGUAGUCCACAGCGAAAGCAAAUCCC--------- ----.......((((((..((..(((.(((.((((((...((((((.....))))))...)))))).))).)))..)).))))))....((....)).......--------- ( -30.60, z-score = -1.10, R) >consensus __________UAGCUAUCACUA_CUUACUCCCUUAUUGUUGCAGGCAAUGUGCUUGUCCAGGCAGGUGGGCAGGGCACAGUAGUCCACUGCGAGGGCAAAUCCC_________ ......................................((((.(((.(((((((((((((......))))))).)))).)).)))....)))).................... (-17.67 = -16.56 + -1.11)

| Location | 7,066,598 – 7,066,693 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 66.83 |
| Shannon entropy | 0.71385 |
| G+C content | 0.52803 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -16.42 |
| Energy contribution | -15.42 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.68 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7066598 95 + 27905053 UGCAGGCUAUGUGCUUGUCCAGGCACGUGGGCAGAGCACAAUAGUCCGCUGCAAGGCUUAUU---CCCAGCAGGGAAGUAAGCAAAAUCAGACACUUA--------- (((((((((((((((((((((......)))))).))))).)))))...)))))..(((((((---(((....))).)))))))...............--------- ( -37.30, z-score = -2.90, R) >droSim1.chr3R 14317697 95 + 27517382 UGCAGGCUAUGUGCUUGUCCAGGCAAGUGGGCAGGGCACAGUAGUCUGCUGCAAGGCUCAAU---CCCAGCAGGGAAGUAAGCAAAAUCAGAGACUUU--------- .((((((((((((((((((((......)))))).)))))).))))))))(((...(((...(---(((....)))))))..)))..............--------- ( -37.80, z-score = -2.42, R) >droSec1.super_0 6228173 95 - 21120651 UGCAGGCUAUGUGCUUGUCCAGGCAAGUGGGCAGGGCACAGUAGUCCGCUGCAAGGCUCAAU---CCCAGCAGGGAAGUAAGCAAAAUCAGAGACUUU--------- .((.(((((((((((((((((......)))))).)))))).))))).))(((...(((...(---(((....)))))))..)))..............--------- ( -33.20, z-score = -1.12, R) >droYak2.chr3R 11099674 95 + 28832112 UGCAGGCAAUGUGCUUGUCCAGGCAAGUGGGCAGAGCACAGUAGUCCGCUGCAAGGCUCAAU---CCCAGCAGGAAACUCAGCAGAAUCAGAGACUUG--------- .((.(((..((((((((((((......)))))).))))))...))).))..((((.(((...---....((((....))..)).......))).))))--------- ( -30.14, z-score = -0.22, R) >droEre2.scaffold_4770 3192883 95 - 17746568 UGCAGGCAAUGUGCUUGUCCAGGCAAGUGGGCAGGGCACAGUAGUCCGCUGCGAGGCUCAAU---CCCAGCAGGGAAGUCAGCAGAAUCAGACACUUG--------- ..((((...((((((((((((......)))))).))))))...(((..((((..((((...(---(((....)))))))).)))).....))).))))--------- ( -34.20, z-score = -0.90, R) >droAna3.scaffold_13340 12930379 95 - 23697760 UGCAGGCAAUGUGCUUGUCCAGGCAGGUGGGCAGAGCACAGUAGUCGACUGCGAAGGCAAUG---CCCAGGAAAGCAGAGACCAGGAUCAGAGACUUC--------- ....(((..(((((((((....))))))(((((..((...((((....))))....))..))---)))......)))..).))...............--------- ( -28.60, z-score = -0.21, R) >dp4.chr2 2203121 98 - 30794189 UGCAGGCCACGUGCUUGUCCAUGCAGGUGGGCAGUGCACAGUAGUCGACGGCGAUGGCAGCU---CCUAGGAUCGCAGCAAGCACAAUGAGAUAAGUCUUC------ (((..(((..((((((((((((....)))))))).))))....((((....))))))).(((---(....))..)).)))........((((....)))).------ ( -29.50, z-score = 0.94, R) >droPer1.super_6 3916895 98 + 6141320 UGCAGGCCACGUGCUUGUCCAUGCAGGUGGGCAGUGCACAGUAGUCGACGGCGAUGGCAGCU---CCUAGGAUCGCAGCAAGCACAAUGAGAUAAGUCUUC------ (((..(((..((((((((((((....)))))))).))))....((((....))))))).(((---(....))..)).)))........((((....)))).------ ( -29.50, z-score = 0.94, R) >droWil1.scaffold_181089 2348077 101 + 12369635 UACAGGCCAAAUUUAAGCCUUGGCAGUGGGGAUUCUGACAAUAGUUUGGAAUUCUUGCCAUUGAAUUUGGUAUGGCGAUGAAGCAAAUAAAAAAUAUAUUU------ ..((.(((((((((((((....)).(..(((((((..(.......)..)))))))..)..))))))))))).))((......)).................------ ( -29.60, z-score = -3.03, R) >droVir3.scaffold_13047 391525 103 - 19223366 UGCAGGCAAUGUGCUUGUCCAAGCACGUGGGCAGAGCACAGUAGUCCACUGCAUUGGCAAUU---UUUGCCAGACCAACUAAUAGAACCAAUACUCCAAUCAUUGU- (((((((..((((((((((((......)))))).))))))...))...)))))((((((...---..)))))).................................- ( -28.90, z-score = -0.96, R) >droMoj3.scaffold_6540 2989019 104 + 34148556 UGCAGGCAAUGUGCUUGUCCAGGCAGGUGGGCAAGGCGCAGUAGUCCACGGAGGCAACGAGG---CCUACUAAGGCUGUGGCCAGCACUAAGAAGCUACAGACUUUG ...((((..((((((((((((......))))))).)))))((.(((......))).))...)---)))..((((((((((((...(.....)..))))))).))))) ( -35.90, z-score = 0.41, R) >droGri2.scaffold_14830 3294272 98 - 6267026 UGCAGGCAAUGUGCUUGUCCAGGCAGGUGGGCAAGGCACAGUAGUCCACAGCGAAAGCAAAU---CCCAAUAGGGCAGUCAGCAGGAGAAAUGCAUUCAUU------ ((((..(..((((((((((((......))))))).)))))....(((...((....))....---(((....))).........))))...))))......------ ( -31.50, z-score = -1.79, R) >consensus UGCAGGCAAUGUGCUUGUCCAGGCAGGUGGGCAGGGCACAGUAGUCCACUGCGAGGGCAAAU___CCCAGCAGGGCAGUAAGCAGAAUCAGAGACUUA_________ (((.(((.(((((((((((((......))))))).)))).)).)))....)))...................................................... (-16.42 = -15.42 + -1.00)
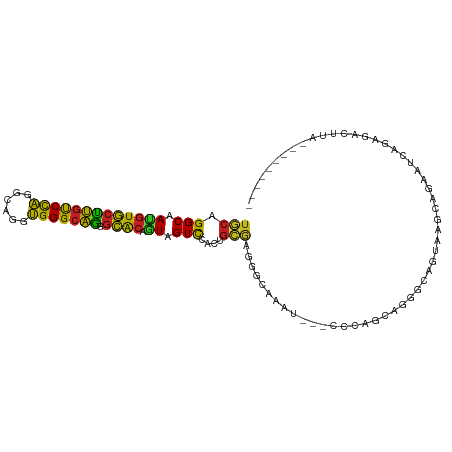
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:35 2011