| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,054,525 – 7,054,634 |
| Length | 109 |
| Max. P | 0.936674 |

| Location | 7,054,525 – 7,054,634 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.51 |
| Shannon entropy | 0.42580 |
| G+C content | 0.54429 |
| Mean single sequence MFE | -41.55 |
| Consensus MFE | -22.29 |
| Energy contribution | -21.64 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7054525 109 + 27905053 ------ACCGCUGGCCGAGUGAAAGACCUCGAAGGAGGUCUGCAGUGUCUUAAUCUCCUGCUCCUUCUCAUCCAGCAGCUGCAGUGACCUCUGGCGCUGCUUCUGCAGUAGCUGU ------...(((((..(((.((.(((((((....)))))))((((.(.........)))))))...)))..)))))((((((((((.((...))))))((....)).)))))).. ( -43.10, z-score = -1.92, R) >droSim1.chr3R 14305663 109 + 27517382 ------ACCGCUGGCCGAGUGAAAGACCUCGAAGGAGGUCUGCAGUGUCUUGAUCUCCUGCUCCUUCUCGUCCAGCAGCUGCAGUGACCUCUGGCGCUGCUUCUGCAGUAGCUGU ------...(((((.((((.((.(((((((....)))))))((((.(.........)))))))...)))).)))))((((((((((.((...))))))((....)).)))))).. ( -46.60, z-score = -2.28, R) >droSec1.super_0 6216362 109 - 21120651 ------ACCGCUGGCCGAGUGAAAGACCUCGAAGGAGGUCUGCAGUGUCUUGAUCUCCUGCUCCUUCUCGUCCAGCAGCUGCAGUGACCUCUGGCGCUGCUUCUGCAGUAGCUGU ------...(((((.((((.((.(((((((....)))))))((((.(.........)))))))...)))).)))))((((((((((.((...))))))((....)).)))))).. ( -46.60, z-score = -2.28, R) >droYak2.chr3R 11087242 109 + 28832112 ------ACCGCUGGCCGAAUGAAAGACCUCGAAGGAGGUCUGCAGUGUCUUGAUCUCCUGCUCCUUCUCAUCAAGAAGCUGCAGUGACCGCUGGCGCUGCUUCUGCAAUAGCUGU ------...((((((.(((....(((((((....)))))))((((((((..(...((((((..(((((.....)))))..)))).))...).))))))))))).))..))))... ( -45.30, z-score = -3.35, R) >droEre2.scaffold_4770 3180640 109 - 17746568 ------ACCGCUGGCCGAAUGGAAGACCUCGAAUGAGGUCUGCAGUGUCUUGAUCUCCUGCUCUUUCUCAUCAAGAAGCUGCAGUGACCGCUGGCGUUGUUUCUGCAAGAGCUGU ------..(((..((.(......(((((((....)))))))(((((.(((((((...............))))))).))))).....).))..)))((((....))))....... ( -36.86, z-score = -0.93, R) >droAna3.scaffold_13340 11615934 109 - 23697760 ------AUUGGUGGCGGAGUGAAAGACCUCGAAAGAGGUCUGCAACGUUUUGAUCUCCUGCUCUUUUUCGUCAAGAAGCUGGAGGGAUCGCUGGCGCUGCUUCUGCAGCAACUGC ------...(.((((((.((...(((((((....)))))))...)).......(((((.((((((.......))).))).)))))..)))))).)(((((....)))))...... ( -45.00, z-score = -2.61, R) >dp4.chr2 28594004 115 + 30794189 GCCGCUACCACCGGCUGUAUGAAAUACCUCAAAUGAGGUCUGCAGCGUCUUGAUCUCCUGCUCCUUCUCGUCGAGAAGCUGCAGGGAUCGCUGUCGCUGCUUCUGCAGCAGCUGC ((.(((.......((((((.(((..(((((....)))))..((((((.(.(((((.(((((..((((((...))))))..))))))))))..).)))))))))))))))))).)) ( -52.71, z-score = -4.71, R) >droPer1.super_6 3930122 115 + 6141320 GCCGCUACCACCGGCUGUAUGAAAUACCUCAAAUGAGGUCUGCAGCGUCUUGAUCUCCUGCUCCUUCUCGUCGAGAAGCUGCAGGGAUCGCUGUCGCUGCUUCUGCAGCAGCUGC ((.(((.......((((((.(((..(((((....)))))..((((((.(.(((((.(((((..((((((...))))))..))))))))))..).)))))))))))))))))).)) ( -52.71, z-score = -4.71, R) >droWil1.scaffold_181130 3309580 112 - 16660200 ---GCCGACUCCCUGGUGAUGGAAGACCUCAAACGAUGUUUUCAAUGUCUUUAUCUCCUGUUCCUUCUCGUCCAAAAGUUGAAGUGAACGUUGUCGCUGUUUCUGCAACAAUUGU ---((((......))))(((.((((((.((....)).))))))...)))..........((((((((.(........)..)))).))))(((((.((.......)).)))))... ( -22.20, z-score = -0.25, R) >droVir3.scaffold_13047 12122940 107 - 19223366 --------UGUUGGGCUUGUGAAACACCUCGAAGGAGGUUUGCAGCGUCUUAAUCUCCUGCUCCUUCUCGUCGAGCAACUGUAAUGAACGCUGCCGCUGCUUUUGGAGCAGCUGC --------.....(((..(..(...(((((....))))))..)(((((..........(((((.........)))))..........))))))))(((((((...)))))))... ( -35.05, z-score = -0.73, R) >droMoj3.scaffold_6540 31162536 107 - 34148556 --------CGUUUGGCUUGUGGAACACCUCGAAGGAGGUCUGCAACGUUUUAAUCUCCUGCUCCUUCUCGUCGAGCAGUUGGAGUGAACGCUGCCGCUGUUUUUGCAGCAACUGC --------.....((((((..((...((((....))))))..)))(((((...(((.((((((.........))))))..)))..)))))..)))(((((....)))))...... ( -38.70, z-score = -1.86, R) >droGri2.scaffold_14830 3298035 107 - 6267026 --------CAUUGGGCUUGUGAAACACUUCGAAGGAGGUUUGCAAUGUUUUAAUCUCCUGCUCCUUCUCGUCGAGCAGCUGCAGCGAUCGUUGCCGCUGCUUCUGCAGCAACUGU --------......(((.(((((.....((((((((((...(((..............))).))))))..))))(((((.(((((....))))).)))))))).)))))...... ( -33.74, z-score = -0.57, R) >consensus ______ACCGCUGGCCGAGUGAAAGACCUCGAAGGAGGUCUGCAGUGUCUUGAUCUCCUGCUCCUUCUCGUCGAGAAGCUGCAGUGACCGCUGGCGCUGCUUCUGCAGCAGCUGU .............(((.........(((((....)))))(((((((...............................)))))))........)))(((((....)))))...... (-22.29 = -21.64 + -0.65)

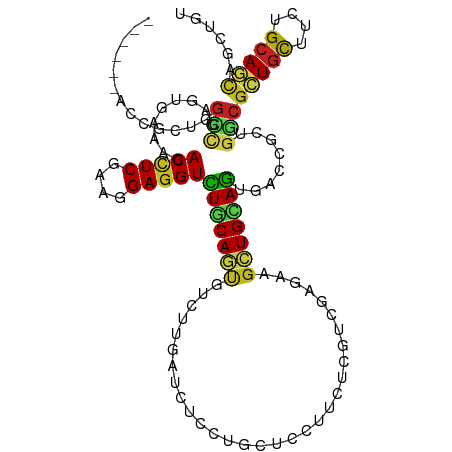

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:32 2011