| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,048,752 – 7,048,856 |
| Length | 104 |
| Max. P | 0.996392 |

| Location | 7,048,752 – 7,048,856 |
|---|---|
| Length | 104 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.89 |
| Shannon entropy | 0.75565 |
| G+C content | 0.36146 |
| Mean single sequence MFE | -18.25 |
| Consensus MFE | -12.15 |
| Energy contribution | -11.75 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.996392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
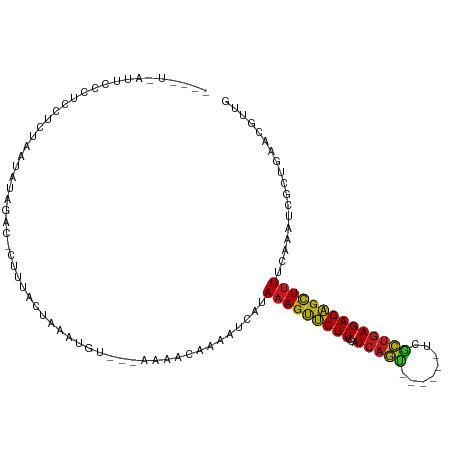
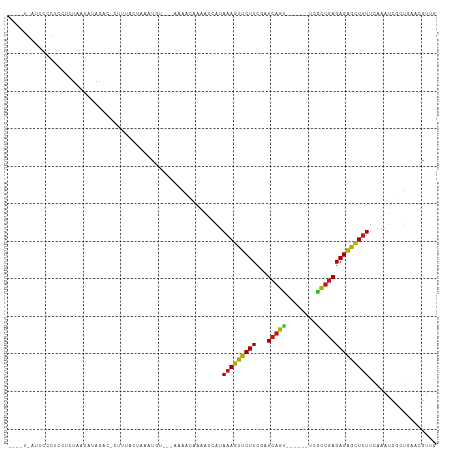
>dm3.chr3R 7048752 104 + 27905053 ACAGUGAUUCCCUCGUGUAAUAUAGACCCC-UAUUAAAUGU---AAAACAAAAUCAUAAAGUUCUUCGAUCAGC------UUGCUGAGAGAGCUUUUCAAAUCGCUAAACGUUG ..(((((((....(((.(((((........-))))).))).---.............(((((((((...((((.------...)))))))))))))...)))))))........ ( -19.10, z-score = -1.86, R) >droAna3.scaffold_13340 11609306 106 - 23697760 ACCUUAAUUUUUUGCUUUUAUUUGAACUCUUAACUUAACGUA--AAAUCCAAACCAUAAAGUUCUUCGAUCAGC------UUGCUGAGAGAGCUUUUAAAAUCGCUGAAUGUUG ....((((.(((.((((((((((((.........)))).)))--)))..........(((((((((...((((.------...))))))))))))).......)).))).)))) ( -14.20, z-score = 0.00, R) >droSim1.chr3R 13197018 105 - 27517382 ACAGUGAUUCCCUCGUAUAAUAGAGACCCCCUAUUAAAUGU---AAAACCAAAUCAUAAAGUUCUUCGAUCAGC------UUGCUGAGAGAGCUUUUCAAAUCGCUGAACGUUG .((((((((....(((.((((((.......)))))).))).---.............(((((((((...((((.------...)))))))))))))...))))))))....... ( -25.30, z-score = -3.75, R) >droSec1.super_0 6210334 105 - 21120651 ACAGUGAUUCCCUCGUGUAAUAGAGCCCCCCUAUUAAAUGU---AAAACAAAAUCAUAAAGUUCUUCGAUCAGC------UUGCUGAGAGAGCUUUUCAAAUCGCUGAACGUUG .((((((((....(((.((((((.......)))))).))).---.............(((((((((...((((.------...)))))))))))))...))))))))....... ( -25.30, z-score = -3.51, R) >droEre2.scaffold_4770 3174831 103 - 17746568 AGAUUGCUUCCCUCAUGUAA--AAGACCCUCUAUUAAAUAU---AAAACAAAAUCAUAAAGUUCUUCGAUCAGC------UUGCUGAGAGAGCUUUUCAAAUCGCUGAACGUUG .((((............(((--.((.....)).))).....---.......))))..(((((((((...((((.------...)))))))))))))(((......)))...... ( -14.65, z-score = 0.09, R) >droYak2.chr3R 11081450 103 + 28832112 AGAGUGCUUCCCUCCUCUAA-AUAACCCCC-UAUUAAAUGU---AAAACAAAAUCAUAAAGUUCUUCGAUCAGC------UUGCUGAGAGAGCUUUUCAAAUCGCUGAACGUUG ((((..........))))..-.........-.....(((((---.............(((((((((...((((.------...)))))))))))))(((......)))))))). ( -15.50, z-score = -0.85, R) >droWil1.scaffold_181130 13356436 94 + 16660200 ----------------AGAAAAUCCAU-CAU-ACUAAAUUAUGCAAAUCAUAA--AUAAAGUUCUUGGAUCAGUGAAAAAUCACUGAGAGAGCUUUUCAAAUCUCUGCUAGUUG ----------------...........-...-....(((((.(((........--..(((((((((...(((((((....)))))))))))))))).........)))))))). ( -20.71, z-score = -2.48, R) >droPer1.super_6 3913881 100 + 6141320 ----AUAGUGUAUUUCCUUGAGAAGAC-CUU-ACUAAAUGCA--AAACUCAUAUCAUAAAGUUCUUCGAUCAGU------ACGCUGAGAGAGCUUUUCAAAUCACUGAACGUCG ----.((((((((((...((((.....-)))-)..)))))).--.............(((((((((...((((.------...))))))))))))).......))))....... ( -18.10, z-score = -0.38, R) >dp4.chr2 28576817 100 + 30794189 ----AUAGUGUAUUUCCUUGAGAAAAC-CUU-ACUAAAUGCA--AAACUCAUAUCAUAAAGUUCUUCGAUCAGU------ACGCUGAGAGAGCUUUUCAAAUCACUGAACGUCG ----.((((((((((...((((.....-)))-)..)))))).--.............(((((((((...((((.------...))))))))))))).......))))....... ( -18.10, z-score = -0.90, R) >droMoj3.scaffold_6540 31156047 93 - 34148556 ----------UGUUCUCAAAGUUUGAU-UUUUACUAACUU----GAAUGAAAAUCAUAAAGUUCUUCGAUCAGU------UCGCUGAGAGAGCUUUUCAAAUCAAUGCAAGUCG ----------.................-........((((----(..(((.......(((((((((...((((.------...))))))))))))).....)))...))))).. ( -17.20, z-score = -0.13, R) >droGri2.scaffold_14830 3288442 97 - 6267026 ----------AAUACAUUUAGACUAAC-UUUAACUAACUCGUACAAACGAAAAACAUAAAGUUCUUCGAUCAGU------UCGCUGAGAGAGCUUUUUAAAUCGAUGCAAGUCG ----------..........((((...-..........((((....)))).......(((((((((...((((.------...))))))))))))).............)))). ( -18.40, z-score = -1.32, R) >droVir3.scaffold_13047 12116467 91 - 19223366 ------------GACCAUUAUUUAGAC-GUUAACUAACUU----AAACGAAACUUAUAAAGUUCUUGGAUCAGU------UCGCUGAGAGAGCUUUUCAAAUCUAUGCAAGUCG ------------(((.......(((((-(((.........----.))))........(((((((((...((((.------...))))))))))))).....)))).....))). ( -16.80, z-score = -0.86, R) >anoGam1.chr2R 41383701 90 + 62725911 --------GGAAUAAAUGUGUAGUGCCUUGCAAUUAAUUA----AUCACCAAAUCGUAAAGCCCUUCGAUCACU---------GCAAGAGGGUUCUAUCAUCGGUUGGUCA--- --------.((.(((...(((((....))))).....)))----.))(((((.(((((.(((((((........---------....))))))).))....))))))))..--- ( -13.90, z-score = 1.18, R) >consensus ____U_AUUCCCUCCUCUAAUAUAGAC_CUUUACUAAAUGU___AAAACAAAAUCAUAAAGUUCUUCGAUCAGU______UCGCUGAGAGAGCUUUUCAAAUCGCUGAACGUUG .........................................................(((((((((...(((((........)))))))))))))).................. (-12.15 = -11.75 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:31 2011