| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,047,890 – 7,047,989 |
| Length | 99 |
| Max. P | 0.759385 |

| Location | 7,047,890 – 7,047,989 |
|---|---|
| Length | 99 |
| Sequences | 15 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.49 |
| Shannon entropy | 0.46257 |
| G+C content | 0.54400 |
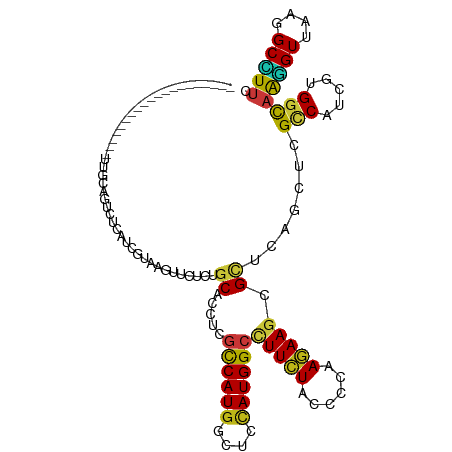
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -16.42 |
| Energy contribution | -15.87 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.759385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
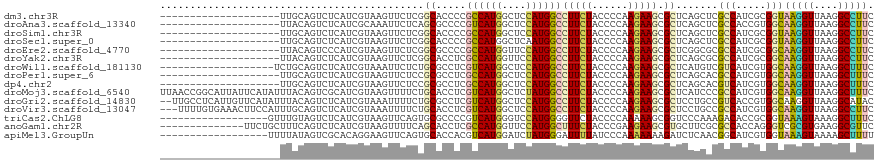
>dm3.chr3R 7047890 99 + 27905053 --------------------UUGCAGUCUCAUCGUAAGUUCUCGGCACCCCGCCAUGGCUCCAUGGCCUUCUACCCCAAGAAGCGCUCAGCUCGCCAUCGCGGUAAGGUUAAGGCCUUC --------------------...............(((..((..((...((((.(((((....(((((((((......))))).)).))....))))).))))....))..))..))). ( -30.90, z-score = -1.64, R) >droAna3.scaffold_13340 11608454 99 - 23697760 --------------------UUACAGUCUCAUCGCAAAUUCUCAGCGCCCCGUCAUGGCUCCAUGGCCUUCUACCCCAAGAAGCGCUCAGCUCGCCACCGUGGCAAGGUUAAGGCCUUC --------------------....................((.(((((...((((((....)))))).((((......))))))))).))...(((.....)))(((((....))))). ( -25.80, z-score = -0.78, R) >droSim1.chr3R 13196154 99 - 27517382 --------------------UUGCAGUCUCAUCGUAAGUUCUCGGCACCCCGCCAUGGCUCCAUGGCCUUCUACCCCAAGAAGCGCUCAGCUCGCCAUCGCGGUAAGGUUAAGGCCUUC --------------------...............(((..((..((...((((.(((((....(((((((((......))))).)).))....))))).))))....))..))..))). ( -30.90, z-score = -1.64, R) >droSec1.super_0 6209470 99 - 21120651 --------------------UUGCAGUCUCAUCGUAAGUUCUCGGCACCCCGCCAUGGCUCAAUGGCCUUCUACCCCAAGAAGCGCUCAGCUCGCCAUCGCGGUAAGGUUAAGGCCUUC --------------------...............(((..((..((...((((.(((((....(((((((((......))))).)).))....))))).))))....))..))..))). ( -30.90, z-score = -1.70, R) >droEre2.scaffold_4770 3174001 99 - 17746568 --------------------UUACAGUCCCAUCGUAAGUUCUCGGCGCCCCGCCAUGGUUCCAUGGCCUUCUACCCCAAGAAGCGCUCGGCGCGCCAUCGCGGCAAGGUUAAGGCCUUC --------------------.....(((.(..(....).....(((((.((((((((....))))))(((((......))))).....)).)))))...).)))(((((....))))). ( -34.20, z-score = -1.54, R) >droYak2.chr3R 11080594 99 + 28832112 --------------------UUACAGUCUCAUCGUAAGUUCUCGGCACCUCGCCAUGGUUCCAUGGCCUUCUACCCCAAGAAGCGCUCAGCGCGCCAUCGCGGCAAGGUUAAGGCCUUC --------------------.......................(((.....((((((....))))))(((..(((.......((((...))))(((.....)))..))).))))))... ( -29.40, z-score = -1.42, R) >droWil1.scaffold_181130 13355628 100 + 16660200 -------------------UCUGCAGUCUCAUCGUAAAUUCUCUGCGCCUCGUCAUGGCUCCAUGGCCUUCUACCCCAAGAAGCGCUCAUGUCGUCAUCGUGGCAAGGUUAAGGCUUUC -------------------...((((................))))(((.((..(((((..(((((((((((......))))).)).))))..))))))).)))(((((....))))). ( -30.19, z-score = -2.82, R) >droPer1.super_6 3913044 99 + 6141320 --------------------UUGCAGUCUCAUCGUAAGUUCUCCGCGCCUCGCCAUGGCUCCAUGGCCUUCUACCCCAAGAAGCGCUCAGCACGCCAUCGUGGCAAGGUUAAGGCUUUC --------------------....(((((.(((....((.((..((((...((((((....)))))).((((......))))))))..)).))(((.....)))..)))..)))))... ( -29.10, z-score = -1.03, R) >dp4.chr2 28575980 99 + 30794189 --------------------UUGCAGUCUCAUCGUAAGUUCUCCGCGCCUCGCCAUGGCUCCAUGGCCUUCUACCCCAAGAAGCGCUCAGCACGUCAUCGUGGCAAGGUUAAGGCUUUC --------------------....(((((.(((...........((((...((((((....)))))).((((......))))))))....((((....))))....)))..)))))... ( -26.80, z-score = -0.63, R) >droMoj3.scaffold_6540 31155254 119 - 34148556 UUAACCGGCAUUAUUCAUAUUUACAGUCGCAUCGUAAGUUUUCUGCACCUCGUCAUGGCUCUAUGGCCUUCUACCCCAAGAAGCGCUCAUCCCGCCAUCGUGGCAAGGUUAAGGCUUUC ((((((.(((........((((((.........))))))....))).((.((..(((((...((((((((((......))))).)).)))...))))))).))...))))))....... ( -28.50, z-score = -1.18, R) >droGri2.scaffold_14830 3287633 117 - 6267026 --UUGCCUCAUUGUUCAUAUUUACAGUCUCAUCGUAAAUUUUCUGCGCCUCGUCAUGGCUCCAUGGCCUUCUACCCCAAGAAGCGCUCCUGCCGUCACCGUGGCAAGGUUAAGGCAUAC --.(((((....((....((((((.........)))))).....))((((.(((((((....((((((((((......))))).......)))))..))))))).))))..)))))... ( -32.01, z-score = -2.25, R) >droVir3.scaffold_13047 12115676 116 - 19223366 ---UUUUGUGAAACUUCCAUUUGCAGUCUCAUCGUAAAUUUUCUGCACCUCGUCAUGGCUCCAUGGCCUUCUACCCCAAGAAGCGCUCCUGCCGCCAUCGUGGCAAGGUUAAGGCCUUC ---....(((((.........(((((................)))))....((((((....)))))).))).)).....((((.(((((((((((....))))).)))....))))))) ( -28.59, z-score = -0.89, R) >triCas2.ChLG8 769889 101 - 15773733 ------------------GUUUGUAGUCUCAUCGUAAGUUCAGUGCGCCCCGUCAUGGGUCCAUGGGGUUCUACCCCAAAAAGCGGUCCCAAAGACACCGCGGUAAAGUAAAGGCUUUC ------------------.................(((((...(((((((......))))((.(((((.....)))))....(((((.........)))))))....)))..))))).. ( -25.60, z-score = -0.52, R) >anoGam1.chr2R 41382763 105 + 62725911 --------------UUCUGCUUUCAGUCUCAUCGUAAGUUUUCAGCACCUCGCCAUGGUUCCAUGGCUUUCUACCCGAAGAAGCGUGCUUCGCGCCACCAGGGUCGCGUGAAGGCGUUC --------------....(((((((..................(((((...((((((....))))))(((((......))))).))))).((((((.....)).))))))))))).... ( -34.90, z-score = -2.12, R) >apiMel3.GroupUn 67141692 101 + 399230636 ------------------UUUUAUAGUCGCACAGGAAGUUCAGUGCACCACGUCAUGGAUCUAUGGGAUUUUAUCCCAAAAAAAGAUCUCAACGGCAUCGUGGUAAAGUAAAAGCUUUU ------------------((((((....((((.((....)).))))((((((.(.(((((((.((((((...)))))).....))))).))...)...))))))...))))))...... ( -25.00, z-score = -2.30, R) >consensus ____________________UUGCAGUCUCAUCGUAAGUUCUCUGCACCUCGCCAUGGCUCCAUGGCCUUCUACCCCAAGAAGCGCUCAGCUCGCCAUCGUGGCAAGGUUAAGGCCUUC ............................................((.....((((((....))))))(((((......))))).)).......(((.....)))(((((....))))). (-16.42 = -15.87 + -0.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:30 2011