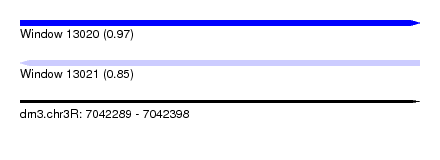
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,042,289 – 7,042,398 |
| Length | 109 |
| Max. P | 0.969981 |

| Location | 7,042,289 – 7,042,398 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 63.23 |
| Shannon entropy | 0.83774 |
| G+C content | 0.38609 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -9.25 |
| Energy contribution | -9.24 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
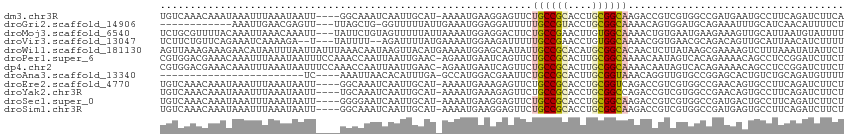
>dm3.chr3R 7042289 109 + 27905053 UGUCAAACAAAUAAAUUUAAAUAAUU----GGCAAAUCAAUUGCAU-AAAAUGAAGGAGUUCUGCCGCACCUGCGGCAAGACCGUCGUGGCCGAUGAAUGCCUUCAGAUCUUCA .(((...........((((..(((((----((....)))))))..)-))).((((((.((..((((((....))))))..))(((((....)))))....)))))))))..... ( -30.80, z-score = -2.54, R) >droGri2.scaffold_14906 2298096 98 - 14172833 ------------AAAUUGAACGAGUU---UUAGCUG-GGUUUUUAUUGAAAUGGAGGAUUUUUGCCGUACCUGCGGCAAAACAGUGGAUGCAGAAAUUUGCAUCAACAUUUUCU ------------........(.(((.---...))).-)(((((....)))))((((((((((((((((....)))))))))..((.(((((((....))))))).))))))))) ( -26.60, z-score = -2.00, R) >droMoj3.scaffold_6540 14833429 111 + 34148556 UCUGCGUUUUACAAAUUAAACAAAUU---UAUUCUGUAGUUUUUAUUAAAAUGGAGGACUUCUGCCGAACUUGUGGCAAAACUGUGAAUGAAGAAAGUUGCAUUAAUGUAUUUU ..(((((((........))))...((---(((((.(.(((((((.........))))))).)(((((......))))).......))))))).......)))............ ( -16.60, z-score = 0.65, R) >droVir3.scaffold_13047 375342 107 - 19223366 UCUUCUGUUCAGAAAUCAAAAGA--U---UAUUUU--AGAUUUUAUGAAAAUGGAAGAUUUUUGCCGAACCUGUGGCAAAACGGUGAACGCAGACAGUUGCAUUAACAUCUUUU ((((((.((((((((((((((..--.---..))))--.)))))).))))...)))))).((((((((......)))))))).((((...((((....)))).....)))).... ( -24.30, z-score = -1.58, R) >droWil1.scaffold_181130 7794992 114 + 16660200 AGUUAAAGAAAGAACAUAAUUUAAUUAUUUAAACAAUAAGUUACAUGAAAAUGGAGCAAUAUUGCCGCACAUGCGGCACAACUCUUAUAAGCGAAAAGUCUUUAAAUAUAUUCU ..(((((((..............................(((.(((....))).))).....((((((....))))))....................)))))))......... ( -20.30, z-score = -1.83, R) >droPer1.super_6 3897227 113 + 6141320 CGUGGACGAAACAAAUUUAAAUAAUUUCCAAACCAAUUAAUUGAAC-AGAAUGAAUCAGUUCUGCCGCACUUGCGGCAAAACAAUAGUCACAGAAAACAGCCUCCGGAUCUUCU .(((.((........(((((.(((((........))))).))))).-...........(((.((((((....)))))).)))....)))))....................... ( -19.90, z-score = -0.80, R) >dp4.chr2 28560233 113 + 30794189 CGUGGACGAAACAAAUUUAAAUAAUUUCCAAACCAAUUAAUUGAAC-AGAAUGAAUCAGUUCUGCCGCACUUGCGGCAAAACAAUAGUCACAGAAAACAGCCUCCGGAUCUUCU .(((.((........(((((.(((((........))))).))))).-...........(((.((((((....)))))).)))....)))))....................... ( -19.90, z-score = -0.80, R) >droAna3.scaffold_13340 11602394 85 - 23697760 ------------------------UC----AAAUUAACACAUUUGA-GCCAUGGACGAAUUCUGCCGCACUUGCGGUAAACAGGUUGUGCCGGAGCACUGUCUGCAGAUGUUUU ------------------------..----........(((((((.-...............((((((....))))))..((((..((((....))))..)))))))))))... ( -24.30, z-score = -0.96, R) >droEre2.scaffold_4770 3168067 109 - 17746568 UGUCAAACAAAUAAAUUUAAAUAAUU----GGCAAAUCAAUUGCAU-AAAAUGAAAGAGUUCUGCCGCACCUGCGGUCAGACCGUCGUGGCCGAACAGUGCCUUCAGAUCUUCU ..........................----((((..(((.((....-.)).)))....((((.(((((....((((.....)))).))))).))))..))))............ ( -24.10, z-score = -1.03, R) >droYak2.chr3R 11074730 109 + 28832112 UGUCAAACAAAUAAAUUUAAAUAAUU----UGCAAAUCAAUUGCAU-AAAAUGAAAGAGUUCUGCCGCACCUGCGGCCAGACCGUCGUGGCCGAACAGUGCCUUCAGAUCUUCU ..........................----(((((.....))))).-.........(((.((((..((((...(((((((.....).))))))....))))...)))).))).. ( -25.50, z-score = -1.76, R) >droSec1.super_0 6203746 109 - 21120651 UGUCAAACAAAUAAAUUUAAAUAAUU----GGGGAAUCAAUUGCAU-AAAAUGAAGGAGUUCUGCCGCACCUGCGGCAAGACCGUCGUGGCCGAUGACUGCCUUCAGAUCUUCU .(((...........((((..(((((----((....)))))))..)-))).((((((.((..((((((....))))))..))(((((....)))))....)))))))))..... ( -31.60, z-score = -2.29, R) >droSim1.chr3R 13190025 109 - 27517382 UGUCAAACAAAUAAAUUUAAAUAAUU----GGCAAAUCAAUUGCAU-AAAAUGAAGGAGUUCUGCCGCACCUGCGGCAAGACCGUCGUGGCCGAUGAGUGCCUUCAGAUCUUCU .(((...........((((..(((((----((....)))))))..)-))).((((((.((..((((((....))))))..))(((((....)))))....)))))))))..... ( -30.80, z-score = -2.40, R) >consensus UGUCAAACAAAUAAAUUUAAAUAAUU____AACCAAUCAAUUGCAU_AAAAUGAAGGAGUUCUGCCGCACCUGCGGCAAAACCGUCGUGGCCGAAAACUGCCUUCAGAUCUUCU ..............................................................((((((....)))))).................................... ( -9.25 = -9.24 + -0.01)

| Location | 7,042,289 – 7,042,398 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 63.23 |
| Shannon entropy | 0.83774 |
| G+C content | 0.38609 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -8.40 |
| Energy contribution | -8.43 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
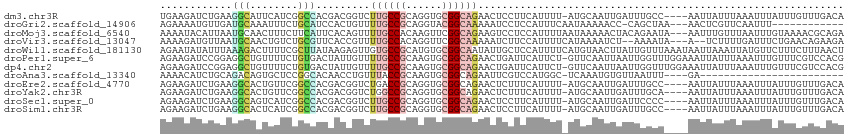
>dm3.chr3R 7042289 109 - 27905053 UGAAGAUCUGAAGGCAUUCAUCGGCCACGACGGUCUUGCCGCAGGUGCGGCAGAACUCCUUCAUUUU-AUGCAAUUGAUUUGCC----AAUUAUUUAAAUUUAUUUGUUUGACA ....((..((((((..(((...((((.....)))).((((((....)))))))))..))))))..))-..((((.....)))).----......((((((......)))))).. ( -29.90, z-score = -1.71, R) >droGri2.scaffold_14906 2298096 98 + 14172833 AGAAAAUGUUGAUGCAAAUUUCUGCAUCCACUGUUUUGCCGCAGGUACGGCAAAAAUCCUCCAUUUCAAUAAAAACC-CAGCUAA---AACUCGUUCAAUUU------------ .((((..((.((((((......)))))).))..((((((((......))))))))........))))..........-.......---..............------------ ( -20.70, z-score = -2.44, R) >droMoj3.scaffold_6540 14833429 111 - 34148556 AAAAUACAUUAAUGCAACUUUCUUCAUUCACAGUUUUGCCACAAGUUCGGCAGAAGUCCUCCAUUUUAAUAAAAACUACAGAAUA---AAUUUGUUUAAUUUGUAAAACGCAGA ............(((..................(((((((........))))))).....................((((((.((---((....)))).))))))....))).. ( -15.80, z-score = -1.03, R) >droVir3.scaffold_13047 375342 107 + 19223366 AAAAGAUGUUAAUGCAACUGUCUGCGUUCACCGUUUUGCCACAGGUUCGGCAAAAAUCUUCCAUUUUCAUAAAAUCU--AAAAUA---A--UCUUUUGAUUUCUGAACAGAAGA (((((((((.((((((......)))))).))..(((((((........)))))))......................--......---)--))))))..(((((....))))). ( -20.10, z-score = -1.17, R) >droWil1.scaffold_181130 7794992 114 - 16660200 AGAAUAUAUUUAAAGACUUUUCGCUUAUAAGAGUUGUGCCGCAUGUGCGGCAAUAUUGCUCCAUUUUCAUGUAACUUAUUGUUUAAAUAAUUAAAUUAUGUUCUUUCUUUAACU (((((((((((((.................((((..((((((....)))))).....))))..............(((((.....)))))))))).)))))))).......... ( -22.80, z-score = -2.00, R) >droPer1.super_6 3897227 113 - 6141320 AGAAGAUCCGGAGGCUGUUUUCUGUGACUAUUGUUUUGCCGCAAGUGCGGCAGAACUGAUUCAUUCU-GUUCAAUUAAUUGGUUUGGAAAUUAUUUAAAUUUGUUUCGUCCACG (((((((((...))..)))))))(((((....((((((((((....)))))))))).((..((....-(((.((.(((((........))))).)).))).))..)))).))). ( -27.10, z-score = -0.87, R) >dp4.chr2 28560233 113 - 30794189 AGAAGAUCCGGAGGCUGUUUUCUGUGACUAUUGUUUUGCCGCAAGUGCGGCAGAACUGAUUCAUUCU-GUUCAAUUAAUUGGUUUGGAAAUUAUUUAAAUUUGUUUCGUCCACG (((((((((...))..)))))))(((((....((((((((((....)))))))))).((..((....-(((.((.(((((........))))).)).))).))..)))).))). ( -27.10, z-score = -0.87, R) >droAna3.scaffold_13340 11602394 85 + 23697760 AAAACAUCUGCAGACAGUGCUCCGGCACAACCUGUUUACCGCAAGUGCGGCAGAAUUCGUCCAUGGC-UCAAAUGUGUUAAUUU----GA------------------------ ......((((((((((((((....))))....)))))..(((....))))))))....((.....))-((((((......))))----))------------------------ ( -20.90, z-score = -0.18, R) >droEre2.scaffold_4770 3168067 109 + 17746568 AGAAGAUCUGAAGGCACUGUUCGGCCACGACGGUCUGACCGCAGGUGCGGCAGAACUCUUUCAUUUU-AUGCAAUUGAUUUGCC----AAUUAUUUAAAUUUAUUUGUUUGACA ....((..((((((..((((.((....))))))((((.((((....))))))))...))))))..))-..((((.....)))).----......((((((......)))))).. ( -24.80, z-score = -0.22, R) >droYak2.chr3R 11074730 109 - 28832112 AGAAGAUCUGAAGGCACUGUUCGGCCACGACGGUCUGGCCGCAGGUGCGGCAGAACUCUUUCAUUUU-AUGCAAUUGAUUUGCA----AAUUAUUUAAAUUUAUUUGUUUGACA .(((((((((...(((((...((((((.(.....)))))))..)))))..))))..)))))......-.(((((.....)))))----......((((((......)))))).. ( -31.10, z-score = -1.87, R) >droSec1.super_0 6203746 109 + 21120651 AGAAGAUCUGAAGGCAGUCAUCGGCCACGACGGUCUUGCCGCAGGUGCGGCAGAACUCCUUCAUUUU-AUGCAAUUGAUUCCCC----AAUUAUUUAAAUUUAUUUGUUUGACA ....((..((((((.((((...((((.....)))).((((((....))))))).)))))))))..))-....(((((......)----))))..((((((......)))))).. ( -28.70, z-score = -1.34, R) >droSim1.chr3R 13190025 109 + 27517382 AGAAGAUCUGAAGGCACUCAUCGGCCACGACGGUCUUGCCGCAGGUGCGGCAGAACUCCUUCAUUUU-AUGCAAUUGAUUUGCC----AAUUAUUUAAAUUUAUUUGUUUGACA ....((..((((((...((...((((.....)))).((((((....))))))))...))))))..))-..((((.....)))).----......((((((......)))))).. ( -28.00, z-score = -1.16, R) >consensus AGAAGAUCUGAAGGCACUUUUCGGCCACGACGGUUUUGCCGCAGGUGCGGCAGAACUCCUUCAUUUU_AUGCAAUUGAUUGGCU____AAUUAUUUAAAUUUAUUUGUUUGACA ............(((........))).........((((((......))))))............................................................. ( -8.40 = -8.43 + 0.03)
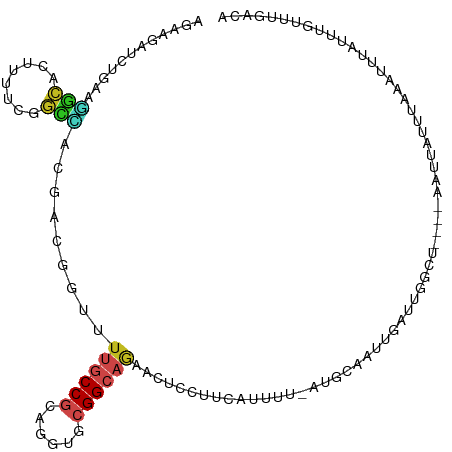
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:05:29 2011